This is a preprint.
UBR-1 enzyme network regulates glutamate homeostasis to affect organismal behavior and developmental viability
- PMID: 40766417
- PMCID: PMC12324394
- DOI: 10.1101/2025.07.28.666006
UBR-1 enzyme network regulates glutamate homeostasis to affect organismal behavior and developmental viability
Abstract
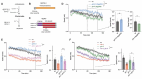
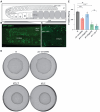
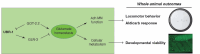
Johanson-Blizzard Syndrome (JBS) is an autosomal recessive spectrum disorder associated with the UBR-1 ubiquitin ligase that features developmental delay including motor abnormalities. Here, we demonstrate that C. elegans UBR-1 regulates high-intensity locomotor behavior and developmental viability via both ubiquitin ligase and scaffolding mechanisms. Super-resolution imaging with CRISPR-engineered UBR-1 and genetic results demonstrated that UBR-1 is expressed and functions in the nervous system including in pre-motor interneurons. To decipher mechanisms of UBR-1 function, we deployed CRISPR-based proteomics using C. elegans which identified a cadre of glutamate metabolic enzymes physically associated with UBR-1 including GLN-3, GOT-2.2, GFAT-1 and GDH-1. Similar to UBR-1, all four glutamate enzymes are genetically linked to human developmental and neurological deficits. Proteomics, multi-gene interaction studies, and pharmacological findings indicated that UBR-1, GLN-3 and GOT-2.2 form a signaling axis that regulates glutamate homeostasis. Developmentally, UBR-1 is expressed in embryos and functions with GLN-3 to regulate viability. Overall, our results suggest UBR-1 is an enzyme hub in a GOT-2.2/UBR-1/GLN-3 axis that maintains glutamate homeostasis required for efficient locomotion and organismal viability. Given the prominent role of glutamate within and outside the nervous system, the UBR-1 glutamate homeostatic network we have identified could contribute to JBS etiology.
Conflict of interest statement
Conflict of Interest The authors declare no competing financial interests.
Figures




References
-
- Zenker M. et al. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome). Nat. Genet. 37, 1345–1350 (2005). - PubMed
-
- Nagashima K., Yagi H. & Kuroume T. A case of Johanson-Blizzard syndrome complicated by diabetes mellitus. Clin. Genet. 43, 98–100 (1993). - PubMed
-
- Al-Dosari M. S. et al. Johanson–Blizzard syndrome: Report of a novel mutation and severe liver involvement. Am. J. Med. Genet. A. 146A, 1875–1879 (2008). - PubMed
Publication types
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
