This is a preprint.
Maternal CENP-C restores centromere symmetry in mammalian zygotes to ensure proper chromosome segregation
- PMID: 40766472
- PMCID: PMC12324218
- DOI: 10.1101/2025.07.23.666394
Maternal CENP-C restores centromere symmetry in mammalian zygotes to ensure proper chromosome segregation
Update in
-
Maternal CENP-C restores centromere symmetry in mammalian zygotes to ensure proper chromosome segregation.Dev Cell. 2026 Jan 14;61(1):146-163.e10. doi: 10.1016/j.devcel.2025.08.017. Epub 2025 Sep 24. Dev Cell. 2026. PMID: 40997799 Free PMC article.
Abstract
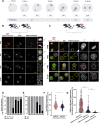
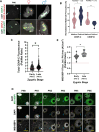
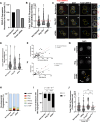
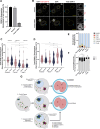
Across metazoan species, the centromere-specific histone variant CENP-A is essential for accurate chromosome segregation, yet its regulation at the parental-to-zygote transition in mammals is poorly understood. To address this, we developed a CENP-A-mScarlet knock-in mouse model, which revealed sex-specific dynamics: mature sperm retains 10% of the CENP-A levels present in MII-oocytes. However, in zygotes prior to the first mitosis, this difference is resolved, using maternally inherited cytoplasmic-CENP-A. Notably, the increase in CENP-A at paternal centromeres is independent of sensing CENP-A asymmetry or the presence of maternal chromosomes. Instead, CENP-A equalization relies on asymmetric recruitment of maternal CENP-C to paternal centromeres. Depletion of maternal CENP-A decreases total CENP-A in pronuclei without disrupting equalization. In contrast, reducing maternal CENP-C or disruption of its dimerization domains impairs CENP-A equalization and chromosome segregation. Therefore, maternal CENP-C acts a key epigenetic regulator that resets centromeric symmetry at fertilization to preserve genome integrity.
Keywords: CENP-A; CENP-C; Centromere; epigenetics; intergenerational; mouse; oocyte; sperm; zygote.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Adenot P.G., Mercier Y., Renard J.P., and Thompson E.M. (1997). Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 124, 4615–4625. - PubMed
-
- Ballow D., Meistrich M.L., Matzuk M., and Rajkovic A. (2006). Sohlh1 is essential for spermatogonial differentiation. Dev. Biol. 294, 161–167. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
