This is a preprint.
Nuclear RNA cap-chaperones eIF4E and NCBP2 govern distinct fates for 1000s of mRNAs uncovering an unexpected regulatory point in gene expression
- PMID: 40766477
- PMCID: PMC12324239
- DOI: 10.1101/2025.07.25.666897
Nuclear RNA cap-chaperones eIF4E and NCBP2 govern distinct fates for 1000s of mRNAs uncovering an unexpected regulatory point in gene expression
Abstract
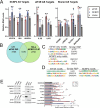
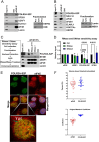
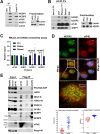
mRNA processing constitutes a series of steps in the nucleus to generate mature mRNAs that can be translated into protein. This relies on the methyl-7-guanosine (m7G) "cap" on the 5'end of mRNAs which is bound by the nuclear cap-binding protein NCBP2 with its cofactor NCBP1. The NCBP1/2 complex chaperones capped mRNA through these processing steps. NCBP2 is considered the sole nuclear cap-binding factor and thus its cap-chaperone role is thought to be a constitutive, housekeeping activity. However, another cap-binding protein, the eukaryotic translation initiation factor eIF4E, is also found in the nucleus. Two cap-binding factors co-existing in the nucleus intimate an undiscovered regulatory point in gene expression or, alternatively, redundancy to ensure gene expression fidelity. Consistent with the former possibility, eIF4E and NCBP2 drove distinct gene expression, transcriptomic, and splicing signatures impacting ~2500 transcripts involved in distinct biological programmes with only ~360 transcripts in common and of these only 79 common splicing events. Thus, each cap-chaperone designates distinct mRNA populations for specific processing revealing a new step in gene expression. We denote this mRNA specification of cap-chaperones (SOCCS). We uncovered multiple molecular mechanisms that contribute to SOCCS: distinct spatial localization of eIF4E and NCBP2 within the nucleus, identification of sequence motifs within targeted mRNAs segregated by eIF4E or NCBP2 sensitivity, distinct protein partners for these cap-chaperones and differential impacts on the production of key spliceosome components e.g. U2AF1, PRP31, SF3B1 and SNRNP200 indicative of distinct transcriptomic landscapes produced by eIF4E or NCBP2 overexpression. In all, the realization that multiple cap-binding proteins coexist in the nucleus led us to identify an unexpected gene-expression regulatory point which engaged distinct biological programmes.
Conflict of interest statement
Disclosing interest statements JCM, CME, BCK, PG, AWBC and KLBB have no disclosure or competing interests. Of note, BCK and KLBB hold patents related to ribavirin use in AML; however, they have received no royalties, compensation or any other financial or other benefit from these patents: Combination therapy using ribavirin as elF4E inhibitor, Inventors: Katherine Borden, Hiba Zahreddine, Biljana Culjkovic-Kraljacic (targeting inducible drug glucuronidation). US10342817B2 GRANTED. Translation dysfunction-based therapeutics Inventors: Gordon Jamieson, Katherine Borden, Biljana Culjkovic, Alex Kentsis: US8497292B2, GRANTED.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
