This is a preprint.
Lower-order methylation states underlie the maintenance and re-establishment of Polycomb modifications in Drosophila embryogenesis
- PMID: 40766521
- PMCID: PMC12324246
- DOI: 10.1101/2025.07.25.666882
Lower-order methylation states underlie the maintenance and re-establishment of Polycomb modifications in Drosophila embryogenesis
Abstract
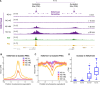
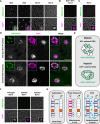
Polycomb Group (PcG) proteins regulate the chromatin composition of an embryo by facilitating the mono, di, and tri-methylation of Histone H3 Lysine 27 (H3K27me1/2/3). For the zygote to inherit an H3K27 methylation blueprint from its mother, PcG-modified states established during oogenesis must persist through early embryogenesis until the onset of large-scale zygotic transcription (Zygotic Genome Activation, ZGA). However, questions have persisted regarding the relative contributions of two molecular mechanisms to the propagation of H3K27 methylation through early development: 1) allosteric regulation of the H3K27 methyltransferase Enhancer of Zeste ( ) by existing H3K27me2/3, and 2) nucleation of activity at chromatin by DNA binding factors. Here, we investigate how allostery and nucleation contribute to H3K27 methylation dynamics in early Drosophila embryogenesis by developing and experimentally validating a mathematical model. This model incorporates measurements of the nuclear concentration dynamics of and the Polycomb Response Element binding factor Pleiohomeotic (Pho), as well as the dilution of epigenetic modifications at DNA replication with the incorporation of histones to nascent chromatin. With stochastic simulations and in vivo experiments, we assert that allosteric regulation of maintains a PcG-imprint on maternal chromosomes in the form of lower-order H3K27 methylation states (H3K27me1/2), that de novo establishment of H3K27 methylation at paternal chromosomes relies on nucleation of activity by Pho, and that broad H3K27me3 domains at both maternal and paternal chromosomes are re-established at ZGA. This work provides a mechanistic explanation for the inheritance of Polycomb states in contexts of intense cellular proliferation.
Figures


Similar articles
-
Nucleation-dependent propagation of Polycomb modifications emerges during the Drosophila maternal to zygotic transition.bioRxiv [Preprint]. 2025 Jul 3:2025.07.02.662854. doi: 10.1101/2025.07.02.662854. bioRxiv. 2025. PMID: 40631074 Free PMC article. Preprint.
-
Polycomb function in early mouse development.Cell Death Differ. 2025 Jan;32(1):90-99. doi: 10.1038/s41418-024-01340-3. Epub 2024 Jul 12. Cell Death Differ. 2025. PMID: 38997437 Review.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Stage-specific DNA methylation dynamics in mammalian heart development.Epigenomics. 2025 Apr;17(5):359-371. doi: 10.1080/17501911.2025.2467024. Epub 2025 Feb 21. Epigenomics. 2025. PMID: 39980349 Review.
-
Flame retardant tetrabromobisphenol A (TBBPA) disrupts histone acetylation during zebrafish maternal-to-zygotic transition.bioRxiv [Preprint]. 2024 Apr 1:2024.03.31.587433. doi: 10.1101/2024.03.31.587433. bioRxiv. 2024. Update in: J Hazard Mater. 2024 Dec 5;480:135845. doi: 10.1016/j.jhazmat.2024.135845. PMID: 38617289 Free PMC article. Updated. Preprint.
References
-
- . Jürgens G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature. 1985. July;316(6024):153–5.
-
- . Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978. Dec;276(5688):565–70. - PubMed
-
- . Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell. 2017. Sept 21;171(1):34–57. - PubMed
-
- . Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007. Jan;8(1):9–22. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
