This is a preprint.
A microsporidial deubiquitinase blocks ubiquitin transfer from adenylated E1 to human UBE2K ubiquitin conjugating enzyme
- PMID: 40766534
- PMCID: PMC12324444
- DOI: 10.1101/2025.07.31.666823
A microsporidial deubiquitinase blocks ubiquitin transfer from adenylated E1 to human UBE2K ubiquitin conjugating enzyme
Abstract
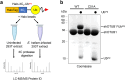
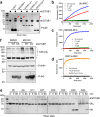
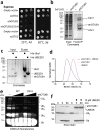
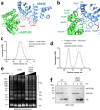
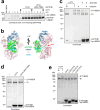
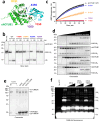
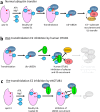
Intracellular pathogens frequently subjugate the ubiquitin system to evade host immune defenses and establish intracellular replication niches. Microsporidia are obligate intracellular animal parasites that typically cause self-limiting infections in humans, but can sometimes cause fatal disseminated disease. At present, the ubiquitin system of microsporidia is virtually unexplored. Here, we discover a likely effector deubiquitinase (DUB) of the otubain subgroup from the human pathogenic microsporidian Encephalitozoon hellem, which we designate ehOTUB1. We find that ehOTUB1 selectively binds the human ubiquitin conjugating (E2) enzyme UBE2K and inhibits its ubiquitin conjugation activity independent of ehOTUB1 DUB activity. We show that ehOTUB1 obstructs docking of UBE2K onto ubiquitin E1 enzyme via steric conflict with ubiquitin in the E1 adenylation site to prevent ubiquitin transfer to UBE2K. This unconventional mechanism of E2 inhibition expands the known repertoire by which pathogens manipulate ubiquitin signaling, and suggests that direct inhibition of E2 enzymes may be a broader function of otubain subfamily DUBs than originally appreciated.
Conflict of interest statement
Competing interests The authors have no competing interests with the content of this work or with the journal.
Figures

Similar articles
-
Microsporidian obligate intracellular parasites subvert autophagy of infected mammalian cells to promote their own growth.mBio. 2025 Jul 9;16(7):e0104925. doi: 10.1128/mbio.01049-25. Epub 2025 May 30. mBio. 2025. PMID: 40444463 Free PMC article.
-
Salmonella exploits host- and bacterial-derived β-alanine for replication inside host macrophages.Elife. 2025 Jun 19;13:RP103714. doi: 10.7554/eLife.103714. Elife. 2025. PMID: 40536105 Free PMC article.
-
Arkadia and Ark2C Promote Substrate Ubiquitylation with Multiple E2 Enzymes.J Mol Biol. 2025 Sep 1;437(17):169259. doi: 10.1016/j.jmb.2025.169259. Epub 2025 May 30. J Mol Biol. 2025. PMID: 40451499
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Direct composite resin fillings versus amalgam fillings for permanent posterior teeth.Cochrane Database Syst Rev. 2021 Aug 13;8(8):CD005620. doi: 10.1002/14651858.CD005620.pub3. Cochrane Database Syst Rev. 2021. PMID: 34387873 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
