This is a preprint.
Multi-omic analyses identify molecular targets of Chd7 that mediate CHARGE syndrome model phenotypes
- PMID: 40766592
- PMCID: PMC12324285
- DOI: 10.1101/2025.07.28.666396
Multi-omic analyses identify molecular targets of Chd7 that mediate CHARGE syndrome model phenotypes
Abstract
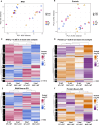
CHARGE syndrome is a developmental disorder that affects 1 in 10,000 births, and patients exhibit both physical and behavioral characteristics. De novo mutations in CHD7 (chromodomain helicase DNA binding protein 7) cause 67% of CHARGE syndrome cases. CHD7 is a DNA-binding chromatin remodeler with thousands of predicted binding sites in the genome, making it challenging to define molecular pathways linking loss of CHD7 to CHARGE phenotypes. To address this problem, here we used a previously characterized zebrafish CHARGE model to generate transcriptomic and proteomic datasets from larval zebrafish head tissue at two developmental time points. By integrating these datasets with differential expression, pathway, and upstream regulator analyses, we identified multiple consistently dysregulated pathways and defined a set of candidate genes that link loss of chd7 with disease-related phenotypes. Finally, to functionally validate the roles of these genes, CRISPR/Cas9-mediated knockdown of capgb, nefla, or rdh5 phenocopies behavioral defects seen in chd7 mutants. Our data provide a resource for further investigation of molecular mediators of CHD7 and a template to reveal functionally relevant therapeutic targets to alleviate specific aspects of CHARGE syndrome.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures





References
-
- Araki K, Meguro H, Kushiya E, et al. (1993) Selective expression of the glutamate receptor channel delta 2 subunit in cerebellar Purkinje cells. Biochemical and biophysical research communications 197(3). Elsevier BV: 1267–1276. - PubMed
-
- Asad Z and Sachidanandan C (2020) Chemical screens in a zebrafish model of CHARGE syndrome identifies small molecules that ameliorate disease-like phenotypes in embryo. European journal of medical genetics 63(2). Elsevier BV: 103661. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
