This is a preprint.
Tranquillyzer: A Flexible Neural Network Framework for Structural Annotation and Demultiplexing of Long-Read Transcriptomes
- PMID: 40766630
- PMCID: PMC12324178
- DOI: 10.1101/2025.07.25.666829
Tranquillyzer: A Flexible Neural Network Framework for Structural Annotation and Demultiplexing of Long-Read Transcriptomes
Abstract
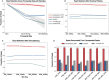
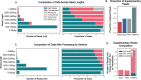
Long-read single-cell RNA sequencing using platforms such as Oxford Nanopore Technologies (ONT) enables full-length transcriptome profiling at single-cell resolution. However, high sequencing error rates, diverse library architectures, and increasing dataset scale introduce major challenges for accurately identifying cell barcodes (CBCs) and unique molecular identifiers (UMIs) - key prerequisites for reliable demultiplexing and deduplication, respectively. Existing pipelines rely on hard-coded heuristics or local transition rules that cannot fully capture this broader structural context and often fail to robustly interpret reads with indel-induced shifts, truncated segments, or non-canonical element ordering. We introduce Tranquillyzer (TRANscript QUantification In Long reads-anaLYZER), a flexible, architecture-aware deep learning framework for processing long-read single-cell RNA-seq data. Tranquillyzer employs a hybrid neural network architecture and a global, context-aware design, and enables precise identification of structural elements - even when elements are shifted, partially degraded, or repeated due to sequencing noise or library construction variability. In addition to supporting established single-cell protocols, Tranquillyzer accommodates custom library formats through rapid, one-time model training on user-defined label schemas, typically completed within a few hours on standard GPUs. Additional features such as scalability across large datasets and comprehensive visualization capabilities further position Tranquillyzer as a flexible and scalable framework solution for processing long-read single-cell transcriptomic datasets.
Keywords: Conditional Random Field; Convolution Neural Network; Long Short-Term Memory; Long-Read; scRNA-seq.
Figures


References
-
- Tian L, Jabbari JS, Thijssen R, Gouil Q, Amarasinghe SL, Voogd O, Kariyawasam H, Du MRM, Schuster J, Wang C, Su S, Dong X, Law CW, Lucattini A, Prawer YDJ, Collar-Fernández C, Chung JD, Naim T, Chan A, Ly CH, Lynch GS, Ryall JG, Anttila CJA, Peng H, Anderson MA, Flensburg C, Majewski I, Roberts AW, Huang DCS, Clark MB, Ritchie ME. Comprehensive characterization of single-cell full-length isoforms in human and mouse with long-read sequencing. Genome Biol 2021;22:310. 10.1186/s13059-021-02525-6. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
