This is a preprint.
Federated Knowledge Retrieval Elevates Large Language Model Performance on Biomedical Benchmarks
- PMID: 40766637
- PMCID: PMC12324469
- DOI: 10.1101/2025.08.01.668022
Federated Knowledge Retrieval Elevates Large Language Model Performance on Biomedical Benchmarks
Abstract
Background: Large language models (LLMs) have significantly advanced natural language processing in biomedical research, however, their reliance on implicit, statistical representations often results in factual inaccuracies or hallucinations, posing significant concerns in high-stakes biomedical contexts.
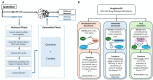
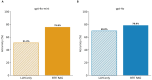
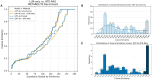
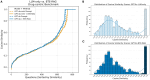
Results: To overcome these limitations, we developed BTE-RAG, a retrieval-augmented generation framework that integrates the reasoning capabilities of advanced language models with explicit mechanistic evidence sourced from BioThings Explorer, an API federation of more than sixty authoritative biomedical knowledge sources. We systematically evaluated BTE-RAG in comparison to traditional LLM-only methods across three benchmark datasets that we created from DrugMechDB. These datasets specifically targeted gene-centric mechanisms (798 questions), metabolite effects (201 questions), and drug-biological process relationships (842 questions). On the gene-centric task, BTE-RAG increased accuracy from 51% to 75.8% for GPT-4o mini and from 69.8% to 78.6% for GPT-4o. In metabolite-focused questions, the proportion of responses with cosine similarity scores of at least 0.90 rose by 82% for GPT-4o mini and 77% for GPT-4o. While overall accuracy was consistent in the drug-biological process benchmark, the retrieval method enhanced response concordance, producing a greater than 10% increase in high-agreement answers (from 129 to 144) using GPT-4o.
Conclusion: Federated knowledge retrieval provides transparent improvements in accuracy for large language models, establishing BTE-RAG as a valuable and practical tool for mechanistic exploration and translational biomedical research.
Conflict of interest statement
Competing Interests The authors declare no competing interests.
Figures
Similar articles
-
Enhancing Pulmonary Disease Prediction Using Large Language Models With Feature Summarization and Hybrid Retrieval-Augmented Generation: Multicenter Methodological Study Based on Radiology Report.J Med Internet Res. 2025 Jun 11;27:e72638. doi: 10.2196/72638. J Med Internet Res. 2025. PMID: 40499132 Free PMC article.
-
Assessing the Accuracy and Reliability of Large Language Models in Psychiatry Using Standardized Multiple-Choice Questions: Cross-Sectional Study.J Med Internet Res. 2025 May 20;27:e69910. doi: 10.2196/69910. J Med Internet Res. 2025. PMID: 40392576 Free PMC article.
-
Role of Artificial Intelligence in Surgical Training by Assessing GPT-4 and GPT-4o on the Japan Surgical Board Examination With Text-Only and Image-Accompanied Questions: Performance Evaluation Study.JMIR Med Educ. 2025 Jul 30;11:e69313. doi: 10.2196/69313. JMIR Med Educ. 2025. PMID: 40737609 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Implementing Large Language Models in Health Care: Clinician-Focused Review With Interactive Guideline.J Med Internet Res. 2025 Jul 11;27:e71916. doi: 10.2196/71916. J Med Internet Res. 2025. PMID: 40644686 Free PMC article. Review.
References
-
- Lin Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science (1979) 379, 1123–1130 (2023). - PubMed
-
- Meier J. et al. Language models enable zero-shot prediction of the effects of mutations on protein function. in Proceedings of the 35th International Conference on Neural Information Processing Systems (Curran Associates Inc., Red Hook, NY, USA, 2021).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
