This is a preprint.
Single Cell RNA Sequencing and Spatial Profiling Identify Mechanisms of Neonatal Brain Hemorrhage Development and Resolution
- PMID: 40766655
- PMCID: PMC12324503
- DOI: 10.1101/2025.07.30.667675
Single Cell RNA Sequencing and Spatial Profiling Identify Mechanisms of Neonatal Brain Hemorrhage Development and Resolution
Abstract
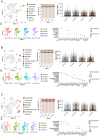
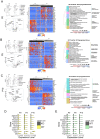
Precise control of cell-cell communication networks within brain neurovascular units (NVUs) promotes normal tissue physiology, and dysregulation of these networks can lead to pathologies including intracerebral hemorrhage (ICH). The cellular and molecular mechanisms underlying ICH development and subsequent tissue repair processes remain poorly understood. Here we employed quantitative single cell RNA sequencing coupled with spatial in situ gene expression profiling to characterize NVU signaling pathways associated with ICH in neonatal mouse brain tissue. The initial stages of ICH pathogenesis are characterized by downregulation of extracellular matrix (ECM)-associated signaling factors (Adamtsl2, Htra3, and Lama4) that functionally connect to canonical TGFβ activation and signaling in vascular endothelial cells. Conversely, the progressive resolution of ICH involves upregulation of neuroinflammatory signaling networks (Gas6 and Axl) alongside activation of iron metabolism pathway components (Hmox1, Cp, and Slc40a1) in astrocytes and microglial cells. Integrated computational modeling identifies additional ligand-receptor signaling networks between perivascular glial cells and endothelial cells during both ICH pathogenesis and resolution. Collectively, these findings illuminate the molecular signaling networks that promote NVU maturation and provide novel mechanistic insights into the pathways controlling ICH pathogenesis and repair.
Conflict of interest statement
COI Disclosure: We have no conflicts of interest to disclose.
Figures








Similar articles
-
Gut microbiota depletion accelerates hematoma resolution and neurological recovery after intracerebral hemorrhage via p-coumaric acid-promoted Treg differentiation.Theranostics. 2025 Jun 9;15(14):6628-6650. doi: 10.7150/thno.113764. eCollection 2025. Theranostics. 2025. PMID: 40585999 Free PMC article.
-
miR-210 Regulates Autophagy Through the AMPK/mTOR Signaling Pathway, Reduces Neuronal Cell Death and Inflammatory Responses, and Enhances Functional Recovery Following Cerebral Hemorrhage in Mice.Neurochem Res. 2025 Jun 5;50(3):180. doi: 10.1007/s11064-025-04434-7. Neurochem Res. 2025. PMID: 40471451 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
From gene to mechanics: a comprehensive insight into the mechanobiology of LMNA mutations in cardiomyopathy.Cell Commun Signal. 2024 Mar 27;22(1):197. doi: 10.1186/s12964-024-01546-5. Cell Commun Signal. 2024. PMID: 38539233 Free PMC article. Review.
-
Immune-mediated disruption of the blood-brain barrier after intracerebral hemorrhage: Insights and potential therapeutic targets.CNS Neurosci Ther. 2024 Jul;30(7):e14853. doi: 10.1111/cns.14853. CNS Neurosci Ther. 2024. PMID: 39034473 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
