WGCNA and single-cell analysis reveal ferroptosis-related gene signatures for hepatocellular carcinoma prognosis and therapy
- PMID: 40767898
- PMCID: PMC12328866
- DOI: 10.1007/s12672-025-03335-z
WGCNA and single-cell analysis reveal ferroptosis-related gene signatures for hepatocellular carcinoma prognosis and therapy
Abstract
Background: Hepatocellular carcinoma (HCC) is a globally serious malignant tumor with high incidence and mortality. Ferroptosis, a newly discovered form of regulated cell death, is significant in tumor initiation and growth. Herein, we performed bioinformatics analysis in order to investigate the expression heterogeneity of ferroptosis-related genes as well as its correlation with HCC clinical outcomes.
Methods: The gene expression data of HCC were downloaded from the TCGA and GEO databases. Weighted Gene Co-expression Network Analysis (WGCNA) was deployed to build gene co-expression networks and explore ferroptosis-related gene modules. We used single-cell RNA sequencing data to analyze the expression of these genes in different cell types. Survival analysis and functional enrichment analysis were utilized to study the biological function and clinical significance of these genes in HCC.
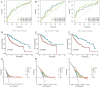
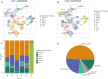
Results: Several ferroptosis-related gene modules were identified by WGCNA, one of which was significantly linked with the clinical characteristics of HCC. Analysis of single-cell sequencing data revealed distinct expression of these core genes in different cell types. Survival analysis revealed that certain ferroptosis-related gene expressions were strongly correlated with patient survival outcomes. Functional enrichment analysis indicated that these genes mainly participate in oxidative stress response, iron metabolism, and apoptosis.
Conclusions: This study reveals the expression heterogeneity of ferroptosis-related genes in HCC and may provide new molecular targets for HCC prognosis and therapy.
Keywords: Bioinformatics; Ferroptosis; Hepatocellular carcinoma.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures












Similar articles
-
Construction of a New Ferroptosis-related Prognosis Model for Survival Prediction in Colorectal Cancer.Curr Med Chem. 2025;32(20):4132-4146. doi: 10.2174/0109298673296767240116215814. Curr Med Chem. 2025. PMID: 38362684
-
Integrative Analysis of Novel Ferroptosis-Related Genes Signatures as Prognostic Biomarkers in Ovarian Cancer.Cancer Rep (Hoboken). 2025 Aug;8(8):e70284. doi: 10.1002/cnr2.70284. Cancer Rep (Hoboken). 2025. PMID: 40744688 Free PMC article.
-
Identification of osteoporosis ferroptosis-related markers and potential therapeutic compounds based on bioinformatics methods and molecular docking technology.BMC Med Genomics. 2024 Apr 22;17(1):99. doi: 10.1186/s12920-024-01872-0. BMC Med Genomics. 2024. PMID: 38650009 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
References
LinkOut - more resources
Full Text Sources
