Identification of copper related biomarkers in breast cancer using machine learning
- PMID: 40767901
- PMCID: PMC12328876
- DOI: 10.1007/s12672-025-03340-2
Identification of copper related biomarkers in breast cancer using machine learning
Abstract
Background: Breast cancer is the most prevalent and deadly cancer among women globally, necessitating more effective diagnostic and therapeutic approaches. This study aims to explore new treatment targets and diagnostic tools.
Methods: Employing machine learning techniques and utilizing PCR, IHC technologies, and multiple databases, we identified and validated genes closely linked with breast cancer and copper-induced cell death. We then explored how their expression levels impact cancer diagnosis, prognosis, immune cell infiltration, and drug sensitivity.
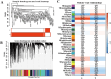
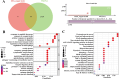
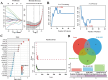
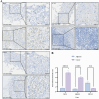
Results: This investigation identified three crucial genes-MT1M, GRHL2, and PKM-intimately associated with the copper death mechanism in breast cancer pathology. Validated through comprehensive analysis across cells, tissue models, and diverse databases, these genes showed significant differential expression (P-value < 0.05), affirming their pivotal role in enhancing diagnostic accuracy (AUC values: 0.917, 0.970, 0.951) and prognostic assessment (HR = 0.65, P = 0.018; HR = 1.69, P = 0.0011; HR = 1.51, P = 0.012) in breast cancer. Additionally, their expression levels influence the infiltration of immune cells and the sensitivity to certain drugs.
Conclusion: MT1M, GRHL2, and PKM are novel diagnostic and therapeutic targets for breast cancer. These findings enhance prognostic evaluations, deepen our understanding of its mechanisms.
Keywords: Biomarker; Breast cancer; Copper death; Immune infiltration; Machine learning.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was conducted using publicly available datasets and cell line experiments. No human participants or animal subjects were involved. Therefore, ethical approval was not required. As this study did not involve any human participants or direct patient interaction, consent to participate was not applicable. Consent for publication: This manuscript does not contain any individual person’s data in any form (including individual details, images, or videos). Therefore, consent to publish is not applicable. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Novel models by machine learning to predict prognosis of breast cancer brain metastases.J Transl Med. 2023 Jun 21;21(1):404. doi: 10.1186/s12967-023-04277-2. J Transl Med. 2023. PMID: 37344847 Free PMC article.
-
Identification of Breast Cancer Subtypes Based on Endoplasmic Reticulum Stress-Related Genes and Analysis of Prognosis and Immune Microenvironment in Breast Cancer Patients.Technol Cancer Res Treat. 2024 Jan-Dec;23:15330338241241484. doi: 10.1177/15330338241241484. Technol Cancer Res Treat. 2024. PMID: 38725284 Free PMC article.
-
Comprehensive pan-cancer analysis reveals NTN1 as an immune infiltrate risk factor and its potential prognostic value in SKCM.Sci Rep. 2025 Jan 25;15(1):3223. doi: 10.1038/s41598-025-85444-x. Sci Rep. 2025. PMID: 39863609 Free PMC article.
-
Cuproptosis: a novel therapeutic mechanism in lung cancer.Cancer Cell Int. 2025 Jun 24;25(1):231. doi: 10.1186/s12935-025-03864-1. Cancer Cell Int. 2025. PMID: 40555995 Free PMC article. Review.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
References
-
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Stat 2021 CA Cancer J Clin. 2021;71:7–33. - PubMed
-
- Libson S, Lippman M. A review of clinical aspects of breast cancer. Int Rev Psychiatry. 2014;26:4–15. - PubMed
-
- Trapani D, Ginsburg O, Fadelu T et al. Global challenges and policy solutions in breast cancer control. Cancer Treatment Reviews [Internet]. 2022 [cited 2023 Aug 29];104. Available from: https://www.cancertreatmentreviews.com/article/S0305-7372(22)00002-0/ful... - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
