Bag-of-words is competitive with sum-of-embeddings language-inspired representations on protein inference
- PMID: 40768408
- PMCID: PMC12327643
- DOI: 10.1371/journal.pone.0325531
Bag-of-words is competitive with sum-of-embeddings language-inspired representations on protein inference
Abstract
Inferring protein function is a fundamental and long-standing problem in biology. Laboratory experiments in this field are often expensive, and therefore large-scale computational protein inference from readily available amino acid sequences is needed to understand in more detail the mechanisms underlying biological processes in living organisms. Recently, studies have utilised mathematical ideas from natural language processing and self-supervised learning, to derive features based on protein sequence information. In the area of language modelling, it has been shown that learnt representations from self-supervised pre-training can capture the semantic information of words well for downstream applications. In this study, we tested the ability of sequence-based protein representations learnt using self-supervised pre-training on a large protein database, on multiple protein inference tasks. We show that simple baseline representations in the form of bag-of-words histograms perform better than those based on self-supervised learning, on sequence similarity and protein inference tasks. By feature selection we show that the top discriminant features help bag-of-words capture important information for data-driven function prediction. These findings could have important implications for self-supervised learning models on protein sequences, and might encourage the consideration of alternative pre-training schemes for learning representations that capture more meaningful biological information from the sequence alone.
Copyright: © 2025 Papadopoulos et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
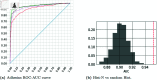
References
-
- Berg JM, Tymoczko JL, Gatto GJJ, Stryer L. Biochemistry. Macmillan; 2015.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

