AND-1 is a critical regulator of R-loop dynamics and a target to overcome endocrine resistance
- PMID: 40768568
- PMCID: PMC12327453
- DOI: 10.1126/sciadv.adv2453
AND-1 is a critical regulator of R-loop dynamics and a target to overcome endocrine resistance
Abstract
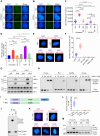
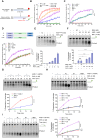
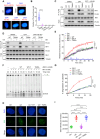
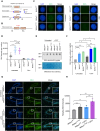
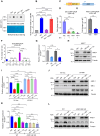
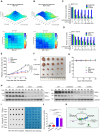
R-loops are three-stranded DNA/RNA hybrids that are essential for various cellular pathways. However, when dysregulated, they lead to genomic instability and numerous human diseases. R-loops are tightly regulated, with RNase H1 acting as a key enzyme responsible for resolving DNA/RNA hybrids. Here, we identify the DNA-binding protein AND-1 as an essential factor in R-loop regulation through directly binding to R-loop structures, where it enhances the recruitment of RNase H1 and stimulates its endonuclease activity. We also provide in vivo evidence that R-loop accumulation occurs in the mammary gland tissue of AND-1-deficient mice. Furthermore, we demonstrate that inhibition of AND-1 decreases ESR1 expression by disrupting R-loop regulation at the enhancer region of the ESR1 gene in estrogen receptor-positive (ER+) breast cancer cells, thereby overcoming resistance to aromatase inhibitors. Collectively, our findings reveal a mechanism by which AND-1 modulates R-loop dynamics and present a promising therapeutic strategy to combat endocrine resistance in breast cancer.
Figures

References
-
- Carles-Kinch K., Kreuzer K. N., RNA-DNA hybrid formation at a bacteriophage T4 replication origin. J. Mol. Biol. 266, 915–926 (1997). - PubMed
-
- Lee D. Y., Clayton D. A., Properties of a primer RNA-DNA hybrid at the mouse mitochondrial DNA leading-strand origin of replication. J. Biol. Chem. 271, 24262–24269 (1996). - PubMed
-
- Yu K., Chedin F., Hsieh C. L., Wilson T. E., Lieber M. R., R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4, 442–451 (2003). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

