A streamlined CRISPR-based test for tuberculosis detection directly from sputum
- PMID: 40768573
- PMCID: PMC12327463
- DOI: 10.1126/sciadv.adx2067
A streamlined CRISPR-based test for tuberculosis detection directly from sputum
Abstract
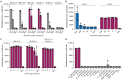
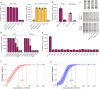
Mycobacterium tuberculosis (Mtb) is a major threat to global health, and there is an urgent need for affordable, simple tuberculosis (TB) diagnosis in underresourced areas. Here, we combine recombinase polymerase amplification with Cas13a and Cas12a detection to create two parallelized one-pot assays that detect two conserved elements of Mtb (IS6110 and IS1081) and a human DNA internal control. These assays are compatible with lateral flow and can be readily lyophilized. Our final assay showed a limit of detection of 69.0 CFU per milliliter for Mtb H37Rv and 80.5 CFU per milliliter for Mycobacterium bovis BCG in spiked sputum, with no cross-reactivity to diverse bacterial or fungal isolates. Clinical tests on 13 blinded sputum samples revealed 100% (six of six) sensitivity and 100% (seven of seven) specificity compared to culture. SHINE-TB streamlines TB diagnosis from sample to answer by combining amplification and detection while being compatible with lateral flow and lyophilization.
Figures


Update of
-
A Streamlined Point-of-Care CRISPR Test for Tuberculosis Detection Directly from Sputum.medRxiv [Preprint]. 2025 Mar 4:2025.02.19.25322517. doi: 10.1101/2025.02.19.25322517. medRxiv. 2025. Update in: Sci Adv. 2025 Aug 8;11(32):eadx2067. doi: 10.1126/sciadv.adx2067. PMID: 40034782 Free PMC article. Updated. Preprint.
References
-
- World Health Organization (WHO), “Global tuberculosis report 2024” (WHO, 2024).
-
- Steingart K. R., Ng V., Henry M., Hopewell P. C., Ramsay A., Cunningham J., Urbanczik R., Perkins M. D., Aziz M. A., Pai M., Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: A systematic review. Lancet Infect. Dis. 6, 664–674 (2006). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

