The cytochrome oxidase defect in ISC-depleted yeast is caused by impaired iron-sulfur cluster maturation of the mitoribosome assembly factor Rsm22
- PMID: 40768618
- PMCID: PMC12375894
- DOI: 10.1002/1873-3468.70129
The cytochrome oxidase defect in ISC-depleted yeast is caused by impaired iron-sulfur cluster maturation of the mitoribosome assembly factor Rsm22
Abstract
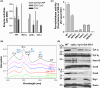
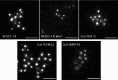
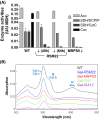
Mitochondria contain the bacteria-inherited iron-sulfur cluster assembly (ISC) machinery to generate cellular iron-sulfur (Fe/S) proteins. Mutations in human ISC genes cause severe disorders with a broad clinical spectrum and are associated with strong defects in mitochondrial Fe/S proteins, including respiratory complexes I-III. For unknown reasons, complex IV (aka cytochrome c oxidase), a non-Fe/S, heme-containing enzyme, is severely affected. Using yeast as a model, we show that depletion of Rsm22, the counterpart of the human mitoribosome assembly factor METTL17, phenocopies the defects observed upon impairing late-acting ISC proteins, that is, diminished activities of mitoribosomal translation and respiratory complexes III and IV. Rsm22 binds Fe/S clusters in vivo, thereby satisfactorily explaining the defect of respiratory complex IV in ISC-deficient cells, because this complex contains three mitochondrial DNA-encoded subunits. Impact statement Defects in mitochondrial Fe/S protein biogenesis also impact respiratory complex IV (COX), even though it lacks Fe/S clusters. Here, we show that the mitoribosome assembly factor Rsm22 binds Fe/S clusters in vivo. Rsm22 maturation defects impair mitoribosomal protein translation including COX subunits, explaining the COX defects in Fe/S cluster-deficient cells.
Keywords: biogenesis; cytochromes; iron–sulfur protein; mitochondrial DNA; mitochondrial ribosomes; respiratory chain complexes; translation.
© 2025 The Author(s). FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

