Microglia-neuron crosstalk through Hex-GM2-MGL2 maintains brain homeostasis
- PMID: 40769205
- PMCID: PMC12545202
- DOI: 10.1038/s41586-025-09477-y
Microglia-neuron crosstalk through Hex-GM2-MGL2 maintains brain homeostasis
Abstract
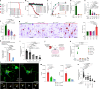
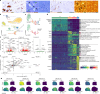
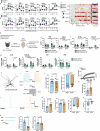
As tissue-resident macrophages of the central nervous system parenchyma, microglia perform diverse essential functions during homeostasis and perturbations1. They primarily interact with neurons by means of synaptic engulfment and through the rapid elimination of apoptotic cells and non-functional synapses2. Here, by combining unbiased lipidomics and high-resolution spatial lipid imaging, deep single-cell transcriptome analysis and novel cell-type-specific mutants, we identified a previously unknown mode of microglial interaction with neurons. During homeostasis, microglia deliver the lysosomal enzyme β-hexosaminidase to neurons for the degradation of the ganglioside GM2 that is integral to maintaining cell membrane organization and function. Absence of Hexb, encoding the β subunit of β-hexosaminidase, in both mice and patients with neurodegenerative Sandhoff disease leads to a massive accumulation of GM2 derivatives in a characteristic spatiotemporal manner3. In mice, neuronal GM2 gangliosides subsequently engage the macrophage galactose-type lectin 2 receptor on microglia through N-acetylgalactosamine residues, leading to lethal neurodegeneration. Notably, replacement of microglia with peripherally derived microglia-like cells is able to break this degenerative cycle and fully restore central nervous system homeostasis. Our results reveal a mode of bidirectional microglia-neuron communication centred around GM2 ganglioside turnover, identify a microgliopathy and offer therapeutic avenues for these maladies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures














References
-
- Prinz, M., Jung, S. & Priller, J. Microglia biology: one century of evolving concepts. Cell179, 292–311 (2019). - PubMed
-
- Sango, K. et al. Mice lacking both subunits of lysosomal β–hexosaminidase display gangliosidosis and mucopolysaccharidosis. Nat. Genet.14, 348–352 (1996). - PubMed
-
- Kierdorf, K., Masuda, T., Jordão, M. J. C. & Prinz, M. Macrophages at CNS interfaces: ontogeny and function in health and disease. Nat. Rev. Neurosci.20, 547–562 (2019). - PubMed
-
- Mrdjen, D. et al. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity48, 380–395 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

