Modulating Antimicrobial Activity and Structure of the Peptide Esc(1-21) via Site-Specific Isopeptide Bond Formation
- PMID: 40769954
- PMCID: PMC12328261
- DOI: 10.1002/psc.70048
Modulating Antimicrobial Activity and Structure of the Peptide Esc(1-21) via Site-Specific Isopeptide Bond Formation
Abstract
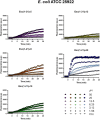
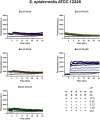
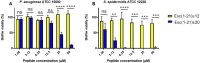
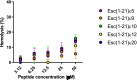
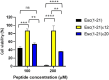
Antimicrobial peptides (AMPs) represent valid alternatives to conventional antibiotics primarily due to their mechanism of action, which consists of cytoplasmic membrane disruption. However, their clinical application is often limited by cytotoxicity at high concentrations and low intrinsic biostability. To address these limitations, various biochemical approaches have been explored. In recent years, the frog-skin derived AMP Esc(1-21) has been extensively characterized for its potent antimicrobial activity, especially against Gram-negative bacteria, both in vitro and in vivo. In this study, we designed and synthesized novel Esc(1-21) analogs in which a single isopeptide bond was introduced in place of a conventional peptide bond at specific positions within the sequence. The resulting five analogs were evaluated for their (i) chemical and structural properties, (ii) resistance to proteolytic degradation, (iii) antimicrobial and antibiofilm activities, (iv) hemolytic and cytotoxic effects, and (v) ability to perturb bacterial cytoplasmic membranes. Among these, Esc(1-21)ε20 showed the most promising features, maintaining antimicrobial and antibiofilm activities comparable to those of the parent peptide while exhibiting lower cytotoxicity towards eukaryotic cells at higher concentrations and greater resistance to enzymatic degradation. These findings highlight Esc(1-21)ε20 as an attractive lead candidate for the development of new antibiotic therapeutics.
© 2025 The Author(s). Journal of Peptide Science published by European Peptide Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
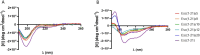
References
-
- Fathi F., Alizadeh B., Tabarzad M. V., and Tabarzad M., “Important Structural Features of Antimicrobial Peptides Towards Specific Activity: Trends in the Development of Efficient Therapeutics,” Bioorganic Chemistry 149 (2024): 107524. - PubMed
-
- Carneri F., Troiano C., Giaquinto G., Roversi D., Franzyk H., and Stella L., “Water‐Membrane Partition and the Mutant Selection Window of Antimicrobial Peptides: Insights From Liposome Studies,” Journal of Colloid and Interface Science 683, no. Pt 1 (2025): 1078–1086. - PubMed
MeSH terms
Substances
Grants and funding
- PRIN2022-2022XFFTH5/Ministero dell'Istruzione, dell'Università e della Ricerca
- PRIN 2020833Y75_005/Ministero dell'Istruzione, dell'Università e della Ricerca
- RG124190CBD34B9D/Sapienza Università di Roma
- ECS 00000024/European Union - NextGenerationEU
- Dr. Barry Sherman Institute for Medicinal Chemistry
LinkOut - more resources
Full Text Sources
Medical
