Aging affects reprogramming of pulmonary capillary endothelial cells after lung injury in male mice
- PMID: 40769983
- PMCID: PMC12328796
- DOI: 10.1038/s41467-025-62431-4
Aging affects reprogramming of pulmonary capillary endothelial cells after lung injury in male mice
Abstract
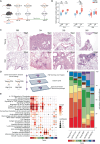
Aging increases the risk of developing fibrotic diseases by hampering tissue regeneration after injury. Using longitudinal single-cell RNA-seq and spatial transcriptomics, here we compare the transcriptome of bleomycin (BLM) -induced fibrotic lungs of young and aged male mice, at 3 time points corresponding to the peak of fibrosis, regeneration, and resolution. We find that lung injury shifts the transcriptomic profiles of three pulmonary capillary endothelial cells (PCEC) subpopulations. The associated signatures are linked to pro-angiogenic signaling with strong Lrg1 expression and do not progress similarly throughout the resolution process between young and old animals. Moreover, part of this set of resolution-associated markers is also detected in PCEC from samples of patients with idiopathic pulmonary fibrosis. Finally, we find that aging also alters the transcriptome of PCEC, which displays typical pro-fibrotic and pro-inflammatory features. We propose that age-associated alterations in specific PCEC subpopulations may interfere with the process of lung progenitor differentiation, thus contributing to the persistent fibrotic process typical of human pathology.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Wijsenbeek, M. & Cottin, V. Spectrum of fibrotic lung diseases. N. Engl. J. Med.383, 958–968 (2020). - PubMed
MeSH terms
Substances
Grants and funding
- ANR-PRCI-18-CE92-0009-01/Agence Nationale de la Recherche (French National Research Agency)
- ANR-22-CE17-0046-01/Agence Nationale de la Recherche (French National Research Agency)
- ANR-15-IDEX-01/Agence Nationale de la Recherche (French National Research Agency)
- ANR-24-CE18-2311/Agence Nationale de la Recherche (French National Research Agency)
- ANR-23-IAHU-0007/Agence Nationale de la Recherche (French National Research Agency)
- ANR-11-LABX-0028-0/Agence Nationale de la Recherche (French National Research Agency)
- BE4443/18-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- BE4443/1-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- BE4443/6-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- KFO309 284237345 P7/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB CRC1213 268555672/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

