A genetically tractable non-vertebrate system to study complete camera-type eye regeneration
- PMID: 40770180
- PMCID: PMC12328594
- DOI: 10.1038/s41467-025-61681-6
A genetically tractable non-vertebrate system to study complete camera-type eye regeneration
Abstract
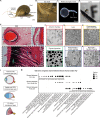
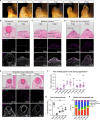
Camera-type eyes are complex sensory organs susceptible to irreversible damage. Their repair is difficult to study due to the paucity of camera-type eye regeneration models. Identifying a genetically tractable organism with the ability to fully regenerate complete camera-type eyes would help overcome this difficulty. Here, we introduce the apple snail Pomacea canaliculata, capable of full regeneration of camera-type eyes even after complete resection. We defined anatomical components of P. canaliculata eyes and genes expressed during crucial steps of their regeneration. By exploiting the unique features of this organism, we successfully established stable mutant lines in apple snails. Our studies reveal that, akin to humans, pax6 is indispensable for eye development in apple snails, establishing this as a research organism to unravel the mechanisms of camera-type eye regeneration. This work expands our understanding of complex sensory organ regeneration and offers a way to explore this process.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Sánchez Alvarado, A. & Tsonis, P. A. Bridging the regeneration gap: genetic insights from diverse animal models. Nat. Rev. Genet.7, 873–884 (2006). - PubMed
-
- Srivastava, M. Beyond Casual Resemblance: Rigorous Frameworks for Comparing Regeneration Across Species. Annu. Rev. Cell Dev. Biol.37, 415–440 (2021). - PubMed
-
- Fernald, R. D. Casting a Genetic Light on the Evolution of Eyes. 313, 1914-1918 (2006). - PubMed
-
- Land, M. F. & Nilsson, D.-E. Animal eyes (Oxford University Press, 2012).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

