GPD1L as a potential biomarker associated with Treg cell infiltration and lipid metabolism in clear cell renal cell carcinoma
- PMID: 40770345
- PMCID: PMC12329951
- DOI: 10.1186/s12967-025-06882-9
GPD1L as a potential biomarker associated with Treg cell infiltration and lipid metabolism in clear cell renal cell carcinoma
Abstract
Background: Clear cell renal cell carcinoma (ccRCC) is a prevalent tumor in the urinary system, presenting a poor prognosis yet being accompanied by a high degree of immune infiltration. Understanding the mechanisms underlying this abnormal infiltration and identifying prognostic biomarkers in this regard is crucial for improving therapeutic outcomes.
Methods: The expression of GPD1L in ccRCC was analyzed using a common database (TCGA). The expression of GPD1L in ccRCC cell lines and tissue samples was verified by western blotting, real time qPCR and immunohistochemistry. The predictive value of GPD1L was evaluated by survival analysis, ROC curve and Cox regression analysis. We used GO, KEGG and gene set enrichment analysis (GSEA) to verify each other. Then the single cell sequencing dataset (GEO) was further analyzed and verified, and the functional phenotype of GPD1L in ccRCC was explored by functional experiments. In addition, the correlation between the expression level of GPD1L and drug resistance of AKT-mTOR pathway was analyzed based on Genomics of Drug Sensitivity in Cancer database (GDSC).
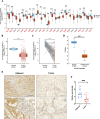
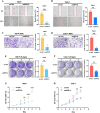
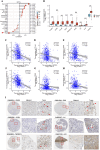
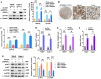
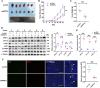
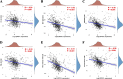
Results: We identified glycerol-3-phosphate dehydrogenase 1-like (GPD1L) as a tumor suppressor gene in ccRCC, and downregulation of GPD1L may facilitate the adaptive survival of tumor cells via enhanced regulatory T cells (Tregs) infiltration and lipid metabolism reprogramming in ccRCC. Our results suggest that there is a significantly diminished GPD1L in ccRCC patients with poorer survival probability. Mechanically, a significant negative correlation between GPD1L expression and Tregs infiltration, and GPD1L-related metabolic analysis reflected the correlation between Tregs and lipid metabolism. In addition, GPD1L expression levels also influence the malignant phenotype of ccRCC and the drug resistance to AKT and mTOR targeted therapy.
Conclusions: Taken together, our results supported GPD1L could be a valuable biomarker for predicting and intervening in ccRCC progression. These insights could shed light on the complex interplay between tumor cell adaptive survival and Treg infiltration, which reflected that the comprehensive and systemic role of GPD1L in ccRCC.
Supplementary Information: The online version contains supplementary material available at 10.1186/s12967-025-06882-9.
Keywords: Biomarker; CcRCC; GPD1L; Immune infiltration; Lipid metabolism; Treg.
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The studies involving human participants were reviewed and approved by The Institutional Research Ethics Committee of Sichuan Provincial People’s Hospital. The patients/participants provided their written informed consent. Consent for publication: All personal data and samples involved in this study have been obtained with their knowledge and permission for publication. Competing interests: The authors declare that they have no competing interests.
Figures



Similar articles
-
Multi-omics analysis of zinc finger protein 683 as a prognostic biomarker for immune infiltration in clear cell renal cell carcinoma.BMC Cancer. 2025 Jul 29;25(1):1236. doi: 10.1186/s12885-025-14643-6. BMC Cancer. 2025. PMID: 40730958 Free PMC article.
-
Computed tomography-based radiomics predicts prognostic and treatment-related levels of immune infiltration in the immune microenvironment of clear cell renal cell carcinoma.BMC Med Imaging. 2025 Jul 1;25(1):213. doi: 10.1186/s12880-025-01749-3. BMC Med Imaging. 2025. PMID: 40596969 Free PMC article.
-
The Role of miR-181a-5p and its Target Genes in the Progression of Clear Cell Renal Cell Carcinoma and Association With the Immune Microenvironment.Clin Genitourin Cancer. 2025 Jun 25:102390. doi: 10.1016/j.clgc.2025.102390. Online ahead of print. Clin Genitourin Cancer. 2025. PMID: 40670255
-
A Systematic Review and Meta-analysis Comparing the Effectiveness and Adverse Effects of Different Systemic Treatments for Non-clear Cell Renal Cell Carcinoma.Eur Urol. 2017 Mar;71(3):426-436. doi: 10.1016/j.eururo.2016.11.020. Epub 2016 Dec 8. Eur Urol. 2017. PMID: 27939075
-
Systemic therapy for non-clear cell renal cell carcinomas: a systematic review and meta-analysis.Eur Urol. 2015 Apr;67(4):740-9. doi: 10.1016/j.eururo.2014.05.010. Epub 2014 Jun 2. Eur Urol. 2015. PMID: 24882670
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

