Primary high-throughput screening of engineered phytases by online monitoring of the oxygen transfer rate of Komagataella phaffii
- PMID: 40770650
- PMCID: PMC12330186
- DOI: 10.1186/s12934-025-02806-w
Primary high-throughput screening of engineered phytases by online monitoring of the oxygen transfer rate of Komagataella phaffii
Abstract
Background: Recombinant phytase production has recently gained increased recognition in phosphate recycling from phytate contained in plant-based side and waste streams. Until now, new phytase variants are evaluated at the end of the expression by standard offline screening procedures, where promising candidates with high activities and protein titers are identified. However, for large mutant libraries, this implies extensive laboratory work for a first screening of hundreds of clones. In this study, for the first time, two synergistic concepts for the primary screening of phytases were investigated.
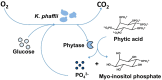
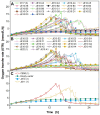
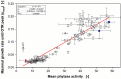
Results: The aim was to predict high recombinant protein producer strains as well as high volumetric activity phytase variants, based on the development of the respiratory activity over time of the host cell, in this case, Komagataella phaffii (Pichia pastoris). In a first step, the metabolic burden was investigated by cultivating a clone library in YPD medium in a µTOM device. It was found that strains expressing medium or high protein concentrations show clear characteristics of an elevated level of metabolic burden during constitutive expression. However, a high protein concentration does not imply a high enzymatic activity. Therefore, in a second approach, the screening was adapted to screen for phytase variants with high volumetric activity. To do so, a modified Syn6 MES medium was developed, where phytic acid was used as the only phosphate source. Thereby, only clones secreting active phytase and generating free phosphate were able to grow, which was monitored via the oxygen transfer rate. A correlation between the offline measured volumetric phytase activity and µmax was found. The clones were then ranked according to their online and offline performance and the results matched in 83% of the cases.
Conclusion: Online monitoring of the oxygen transfer rates in 96-well plates allowed for the evaluation of the total protein concentration and the volumetric phytase activity already during the expression. Using these results, also the specific activity can be calculated. In the future, primary screening experiments of large enzyme mutant libraries can be conducted without offline activity assays, to identify promising candidates.
Keywords: Komagataella phaffii; Pichia pastoris; Phytase screening; Recombinant protein production; Volumetric and specific enzyme activity.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Systemic Inflammatory Response Syndrome.2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31613449 Free Books & Documents.
-
[Volume and health outcomes: evidence from systematic reviews and from evaluation of Italian hospital data].Epidemiol Prev. 2013 Mar-Jun;37(2-3 Suppl 2):1-100. Epidemiol Prev. 2013. PMID: 23851286 Italian.
-
Single-Stage Pedicle Preputial Tube Substitution Urethroplasty with Corpora Cavernosa Augmentation Using Buccal Mucosa Graft for Primary Peno-Scrotal Hypospadias Re-pair in Adults.Int Braz J Urol. 2025 Jul-Aug;51(4):e20240650. doi: 10.1590/S1677-5538.IBJU.2024.0650. Int Braz J Urol. 2025. PMID: 40105707 Free PMC article.
References
-
- Rittmann BE, Mayer B, Westerhoff P, Edwards M. Capturing the lost phosphorus. Chemosphere. 2011;84(6):846–53. - PubMed
-
- Weiner M, Salminen W, Larson P, Barter R, Kranetz J, Simon G. Toxicological review of inorganic phosphates. Food Chem Toxicol. 2001;39(8):759–86. - PubMed
-
- Chandrajith R, Dissanayake C. Phosphate mineral fertilizers, trace metals and human health. J Natl Sci Foundation Sri Lanka. 2009;37(3):153-165.
-
- Killiches F, Phosphat. Mineralischer Rohstoff und unverzichtbarer Nährstoff für die Ernährungssicherheit weltweit. 2013 [Available from: https://www.bgr.bund.de/DE/Themen/Zusammenarbeit/TechnZusammenarbeit/Pol... Accessed 26.11.2024.
-
- Schnug E, Lottermoser BG. Fertilizer-derived uranium and its threat to human health. Environ Sci Technol. 2013;47(6):2433–4. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

