Multi-omics profiling identifies TNFRSF18 as a novel marker of exhausted CD8⁺ T cells and reveals tumour-immune dynamics in colorectal cancer
- PMID: 40770837
- PMCID: PMC12328248
- DOI: 10.1002/ctm2.70425
Multi-omics profiling identifies TNFRSF18 as a novel marker of exhausted CD8⁺ T cells and reveals tumour-immune dynamics in colorectal cancer
Abstract
Background: Colorectal cancer (CRC) ranks among the most prevalent malignant tumours of the digestive system globally and is associated with unfavourable survival outcomes. The exhaustion of CD8⁺ T cells serves a crucial role in facilitating tumour immune escape. Yet, the dynamic evolution of CD8⁺ T cell exhaustion and its impact on clinical prognosis across TNM (tumour-node-metastasis) stages in CRC remains incompletely characterized.
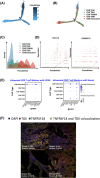
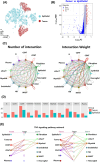
Methods: Tumour and adjacent tissues (20 samples total) from 6 CRC patients spanning diverse TNM stages were analyzed using integrated single-cell transcriptomic profiling (scRNA-seq), single-cell T cell receptor/B cell receptor sequencing (scVDJ-seq), and spatial transcriptomics. T cell exhaustion markers, immune clonality, gene expression profiles, and the spatial distribution of both tumour cells and immune cells were systematically profiled. Functional enrichment and intercellular communication analyses were conducted. Key findings were validated using immunofluorescence and public datasets.
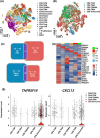
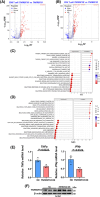
Results: Our results illustrate how advancing TNM stages in CRC shape CD8⁺ T cell exhaustion through divergent TNFRSF18/CXCL13 dynamics and ribosomal stemness. TNFRSF18 expression was notably higher in T cells infiltrating tumour tissues relative to their counterparts in adjacent non-tumorous areas, with high-expressing CD8⁺ T cells exhibiting marked exhaustion features. During CRC progression, TNM-stage-driven remodelling of the tumour microenvironment (TME) induced progressive CD8⁺ T cell exhaustion marked by declining TNFRSF18 and rising CXCL13 expression in tumour-infiltrating T cells elevation of both markers in the tumour compared with adjacent tissues. Moreover, we show that tumour cells displayed elevated expression of stemness-associated ribosomal genes (RPS7, RPL8, RPL30), peaking at stage T4, which correlated with poor prognosis and immune escape.
Conclusions: This integrative multi-omics study uncovers CD8⁺ T cell exhaustion dynamics and ribosomal stemness-mediated immune evasion across CRC progression. CXCL13, TNFRSF18, and ribosomal proteins (RPS7/RPL8/RPL30) are identified as novel biomarkers with direct prognostic value and therapeutic relevance, providing therapeutic targets for precision immunotherapy in CRC.
Key points: Multi-omics analysis reveals dynamic CD8+ T cell exhaustion patterns across CRC samples with different TNM stages. TNFRSF18 is highly expressed in exhausted tumour-infiltrating CD8+ T cells and declines with disease progression. Ribosomal stemness in tumour cells promotes immune evasion by impairing TNF-mediated CD8+ T cell function.
Keywords: T cell exhaustion; TNFRSF18 (GITR); TNM stage; colorectal cancer; singlecell RNA sequencing; spatial transcriptomics.
© 2025 The Author(s). Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Interplay between tumor mutation burden and the tumor microenvironment predicts the prognosis of pan-cancer anti-PD-1/PD-L1 therapy.Front Immunol. 2025 Jul 24;16:1557461. doi: 10.3389/fimmu.2025.1557461. eCollection 2025. Front Immunol. 2025. PMID: 40777041 Free PMC article.
-
Multi-omics analysis unveils the predictive value of IGF2BP3/SPHK1 signaling in cancer stem cells for prognosis and immunotherapeutic response in muscle-invasive bladder cancer.J Transl Med. 2024 Oct 4;22(1):900. doi: 10.1186/s12967-024-05685-8. J Transl Med. 2024. PMID: 39367493 Free PMC article.
-
Diversity of mast cell subpopulations in the tumor microenvironment of colorectal cancer and their prognostic implications.Cancer Immunol Immunother. 2025 Jun 30;74(8):255. doi: 10.1007/s00262-025-04119-8. Cancer Immunol Immunother. 2025. PMID: 40586903 Free PMC article.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
The role of intratumoral microbiome in the occurrence, proliferation, metastasis of colorectal cancer and its underlying therapeutic strategies.Ageing Res Rev. 2025 Sep;111:102820. doi: 10.1016/j.arr.2025.102820. Epub 2025 Jul 8. Ageing Res Rev. 2025. PMID: 40639623 Review.
References
MeSH terms
Substances
Grants and funding
- 21KJA310002/Natural Science Foundation of Jiangsu Higher Education Institution-Key Program
- Q424900220/Soochow University Research Development Funds
- 31470840/National Natural Science Foundation of China
- SLJ2022011/Suzhou Technology Bureau
- NCTIB2024JS0101/Provincial-level Talent Program for National Center of Technology Innovation for Biopharmaceuticals
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
