CRISPR RiPCA for Investigating eIF4E-m7GpppX Capped mRNA Interactions
- PMID: 40770919
- PMCID: PMC12409724
- DOI: 10.1021/acschembio.5c00471
CRISPR RiPCA for Investigating eIF4E-m7GpppX Capped mRNA Interactions
Abstract
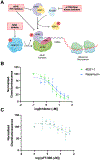
Post-transcriptional modifications expand the information encoded by an mRNA. These dynamic and reversible modifications are specifically recognized by reader RNA-binding proteins (RBPs), which mediate the regulation of gene expression, RNA processing, localization, stability, and translation. Given their crucial functions, any disruptions in the normal activity of these readers can have significant implications for cellular health. Consequently, the dysregulation of these RBPs has been associated with neurodegenerative disorders, cancers, and viral infections. Therefore, there has been growing interest in targeting reader RBPs as a potential therapeutic strategy since developing molecules that restore proper RNA processing and function may offer a promising avenue for treating diseases. In this work, we coupled our previously established live-cell RNA-protein interaction (RPI) assay, RNA interaction with Protein-mediated Complementation Assay (RiPCA), with CRISPR technology to build a new platform, CRISPR RiPCA. As a model for development, we utilized the interaction of eukaryotic translation initiation factor 4E (eIF4E), a reader RBP that binds to the m7GpppX cap present at the 5' terminus of coding mRNAs, with an m7G capped RNA substrate. Using eIF4E CRISPR RiPCA, we demonstrate our technology's potential for measuring on-target activity of inhibitors of the eIF4E RPI of relevance to cancer drug discovery.
Figures




Update of
-
CRISPR RiPCA for Investigating eIF4E-m7GpppX Capped mRNA Interactions.bioRxiv [Preprint]. 2025 Jun 22:2025.06.19.660603. doi: 10.1101/2025.06.19.660603. bioRxiv. 2025. Update in: ACS Chem Biol. 2025 Aug 15;20(8):2038-2048. doi: 10.1021/acschembio.5c00471. PMID: 40666878 Free PMC article. Updated. Preprint.
Similar articles
-
CRISPR RiPCA for Investigating eIF4E-m7GpppX Capped mRNA Interactions.bioRxiv [Preprint]. 2025 Jun 22:2025.06.19.660603. doi: 10.1101/2025.06.19.660603. bioRxiv. 2025. Update in: ACS Chem Biol. 2025 Aug 15;20(8):2038-2048. doi: 10.1021/acschembio.5c00471. PMID: 40666878 Free PMC article. Updated. Preprint.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Biochemical analysis of human eIF4E-DCP2 interaction: Implications for the relationship between translation initiation and decapping.PLoS One. 2025 Aug 1;20(8):e0322271. doi: 10.1371/journal.pone.0322271. eCollection 2025. PLoS One. 2025. PMID: 40748882 Free PMC article.
-
Emerging Roles of m7G-Cap Hypermethylation and Nuclear Cap-Binding Proteins in Bypassing Suppression of eIF4E-Dependent Translation.Viruses. 2025 Mar 5;17(3):372. doi: 10.3390/v17030372. Viruses. 2025. PMID: 40143300 Free PMC article. Review.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
References
-
- Delaunay S; Helm M; Frye M RNA Modifications in Physiology and Disease: Towards Clinical Applications. Nat. Rev. Genet 2024, 25 (2), 104–122. - PubMed
-
- Barbieri I; Kouzarides T Role of RNA Modifications in Cancer. Nat. Rev. Cancer 2020, 20 (6), 303–322. - PubMed
-
- Matsumura Y; Wei F-Y; Sakai J Epitranscriptomics in Metabolic Disease. Nat. Metab 2023, 5 (3), 370–384. - PubMed
-
- Soueid DM; Garner AL RNA-Protein Interactions: A New Approach for Drugging RNA Biology. In Methods and Principles in Medicinal Chemistry; Schneekloth J, Pettersson M, Mannhold R, Buschmann H, Holenz J, Eds.; Wiley, 2024; pp 281–319, DOI: 10.1002/9783527840458.ch11. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

