Genome-wide association study reveals the genetic basis of vitamin C content in rapeseed (Brassica napus L.) seedlings
- PMID: 40772053
- PMCID: PMC12325403
- DOI: 10.3389/fpls.2025.1649023
Genome-wide association study reveals the genetic basis of vitamin C content in rapeseed (Brassica napus L.) seedlings
Abstract
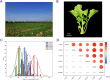
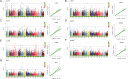
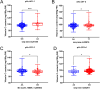
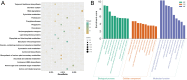
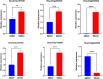
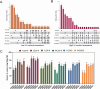
Rapeseed (Brassica napus L.) is a versatile crop, with its seedlings and flowering stalks can be utilized as vegetables, which are rich in vitamin C (Vc) and other essential nutrients, including selenium, calcium, zinc, and various amino acids. Despite the well-documented health benefits of Vc as a critical antioxidant nutrient, the genetic mechanisms governing Vc accumulation in rapeseed remain poorly understood. In this study, we investigated the Vc content of 327 rapeseed accessions during the seedling stage across six environments in Hubei province over three consecutive years (2018-2020). The Vc content in these environments ranged from 62.82 to 161.25 mg/100g, demonstrating high genetic variation (7.96% to 9.43%) and heritability (86.11%). Genome-wide association studies (GWAS) identified 31 significant single nucleotide polymorphisms (SNPs) across various chromosomes, which explained 5.68% to 12.78% of the phenotypic variation, integrated into 16 quantitative trait loci (QTLs). Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses of the 2365 annotated genes near associated SNPs revealed significant involvement in diverse metabolic pathways including peroxisome, ascorbate, secondary metabolites, and terpenoid biosynthesis, as well as biological processes such as hydrogen peroxide/lactate metabolism and ROS biosynthesis, along with associations with specific cellular components and molecular functions. Furthermore, we identified six candidate genes that exhibit significant differences in expression between low and high Vc accessions, which are potentially involved in Vc biosynthesis but require further experimental validation. Additionally, we selected four superior germplasms (8S079, 8S200, 8S242, and 8S243) that demonstrate extreme Vc content, providing valuable germplasm resources for breeding. Collectively, these findings provide novel and comprehensive insights into the genetic and molecular mechanisms regulating Vc accumulation in rapeseed, thereby establishing a foundation for targeted genetic improvement of nutritional quality in vegetable rapeseed varieties.
Keywords: GWAS; candidate genes; germplasm resource; rapeseed seedlings; vitamin C.
Copyright © 2025 Wang, Kuang, Tian, Wang, Wang and Dun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

