Genome-wide identification of GATA family genes in sweet potato (Ipomoea batatas L.) and their expression patterns under abiotic stress
- PMID: 40772271
- PMCID: PMC12326740
- DOI: 10.3389/fgene.2025.1635749
Genome-wide identification of GATA family genes in sweet potato (Ipomoea batatas L.) and their expression patterns under abiotic stress
Abstract
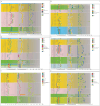
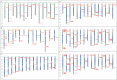
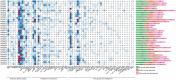
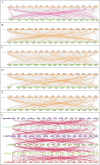
GATAs, a type of zinc finger protein transcription factors, can bind to DNA regulatory regions to control the expression of target genes, thereby affecting plant growth and development under normal conditions or environmental stress. However, the GATA gene family has not been identified in sweet potato. In this study, a total of 35, 33, 34, 39, 63, and 56 GATA genes were identified in sweet potato, Ipomoea aquatica, Ipomoea cairica, Ipomoea nil, Ipomoea triloba, and Ipomoea trifida, respectively. Phylogenetic analysis categorized the GATA genes into six groups according to their distinct features, and this classification was validated by the structural characteristics of exons/introns and conserved motif analysis. The cis-acting elements located in the promoter regions were also found to be enriched with biotic and abiotic responsive elements, which may play a pivotal role in plant stress adaptation. Then the gene duplication events and synteny between the genome of sweet potato and those of Ipomoea aquatica, Ipomoea cairica, Ipomoea nil, Ipomoea triloba, and Ipomoea trifida were analyzed, which provided insights into evolutionary mechanisms. Moreover, expression pattern analysis was performed on IbGATA genes, many of which were significantly induced by multiple types of abiotic stress, which may render these genes candidates for molecular breeding strategies in sweet potato. Overall, this experiment conducted a systematic exploration of GATA genes by investigating their evolutionary relationships, structural characteristics, functional properties, and expression patterns, thereby establishing a theoretical foundation for further in-depth research on the features of the GATA gene family.
Keywords: GATA transcription factor; abiotic stress; drought and salt stress; genome-wide; sweet potato (Ipomoea batatas L.).
Copyright © 2025 Wang, Lan, Xiao, Peng, Pan, Deng and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Chen C., Zhang K. X., Khurshid M., Li J. B., He M., Georgiev M. I., et al. (2019). MYB transcription repressors regulate plant secondary metabolism. Crit. Rev. Plant Sci. 38 (3), 159–170. 10.1080/07352689.2019.1632542 - DOI
-
- Chen F., Hu Y., Vannozzi A., Wu K. C., Cai H. Y., Qin Y., et al. (2018). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36 (5-6), 311–335. 10.1080/07352689.2018.1441103 - DOI
LinkOut - more resources
Full Text Sources

