Circular RNA Profiling Identifies circ5078 as a BMPR2-Derived Regulator of Endothelial Proliferation and Stress Responses
- PMID: 40772817
- PMCID: PMC12379793
- DOI: 10.1161/ATVBAHA.124.322455
Circular RNA Profiling Identifies circ5078 as a BMPR2-Derived Regulator of Endothelial Proliferation and Stress Responses
Abstract
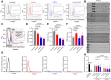
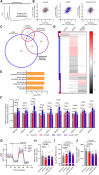
Background: The BMPR2 gene encodes the BMPR-II (bone morphogenetic protein receptor type-II) and is a known regulator of endothelial proliferation, apoptosis, and translational stress responses. While these effects are generally attributed to the actions of BMPR-II protein, we used circular RNA profiling to identify circ3218 and circ5078 as new BMPR2-derived functional RNAs.
Methods: Circular RNAs were profiled by ultradeep RNA sequencing of human pulmonary artery endothelial cells. Novel BMPR2-derived circular RNAs were assessed for their effects on endothelial proliferation, apoptosis, and translational stress responses in human pulmonary artery endothelial cells and endothelial cells from patients with pulmonary arterial hypertension with heterozygous loss-of-function BMPR2 mutations. circ5078 to linear BMPR2 mRNA ratios were quantified in cultured lymphocytes from patients with pulmonary arterial hypertension with BMPR2 mutations versus unaffected mutation carriers.
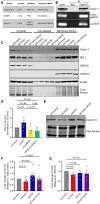
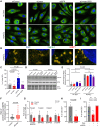
Results: Depletion of circ3218 enhanced human pulmonary artery endothelial cell apoptosis, whereas circ5078 silencing increased proliferation. Enhanced proliferation with circ5078 depletion was eliminated by cosilencing of circ5078 with either linear BMPR2 mRNA or the stress granule protein, Caprin-1, indicating a potential interdependence of BMPR2 transcripts in the regulation of endothelial function. Patients with pulmonary arterial hypertension with BMPR2 mutations exhibited increased circ5078 to linear BMPR2 mRNA ratios, alongside impaired stress responses in patient endothelial cells that were deficient in linear BMPR2 transcripts but not circ5078. circ5078 depletion enhanced stress responses in human pulmonary artery endothelial cells and rescued stress granule formation in patient endothelial cells, independent of BMPR-II protein levels. Assessment of translational regulation by polysome profiling did not identify any impact of linear or circular BMPR2 transcript loss on global protein synthesis or stress-induced eIF2α (eukaryotic initiation factor 2α) phosphorylation but did identify the enhanced translational efficiency of select nuclear-encoded mitochondrial ribosome proteins with circ5078 depletion, offering a link between mitochondrial function and the circ5078-deficient endothelial phenotype.
Conclusions: The identification of circ3218 and circ5078 as novel BMPR2-derived gene products reveals interdependent roles for coding and noncoding BMPR2 transcripts as regulators of endothelial function.
Keywords: RNA, circular; endothelial cells; phosphorylation; pulmonary arterial hypertension; stress granules.
Conflict of interest statement
None.
Figures



Comment in
-
Circular RNAs Complement Cardiovascular Messaging Machinery.Arterioscler Thromb Vasc Biol. 2025 Sep;45(9):1562-1564. doi: 10.1161/ATVBAHA.125.323371. Epub 2025 Aug 7. Arterioscler Thromb Vasc Biol. 2025. PMID: 40772291 No abstract available.
References
-
- Toshner M, Voswinckel R, Southwood M, Al-Lamki R, Howard LS, Marchesan D, Yang J, Suntharalingam J, Soon E, Exley A, et al. Evidence of dysfunction of endothelial progenitors in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2009;180:780–787. doi: 10.1164/rccm.200810-1662OC - PMC - PubMed
-
- Sawada H, Saito T, Nickel NP, Alastalo TP, Glotzbach JP, Chan R, Haghighat L, Fuchs G, Januszyk M, Cao A, et al. Reduced BMPR2 expression induces GM-CSF translation and macrophage recruitment in humans and mice to exacerbate pulmonary hypertension. J Exp Med. 2014;211:263–280. doi: 10.1084/jem.20111741 - PMC - PubMed
-
- Machado RD, Aldred MA, James V, Harrison RE, Patel B, Schwalbe EC, Gruenig E, Janssen B, Koehler R, Seeger W, et al. Mutations of the TGF-beta type II receptor BMPR2 in pulmonary arterial hypertension. Hum Mutat. 2006;27:121–132. doi: 10.1002/humu.20285 - PubMed
-
- Cober ND, VandenBroek MM, Ormiston ML, Stewart DJ. Evolving concepts in endothelial pathobiology of pulmonary arterial hypertension. Hypertension. 2022;79:1580–1590. doi: 10.1161/HYPERTENSIONAHA.122.18261 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

