Reduction of Tooth Replacement Disproportionately Affects the Evolution of Enamel Matrix Proteins
- PMID: 40773061
- PMCID: PMC12354546
- DOI: 10.1007/s00239-025-10258-4
Reduction of Tooth Replacement Disproportionately Affects the Evolution of Enamel Matrix Proteins
Abstract
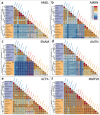
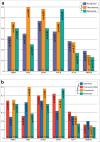
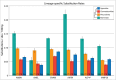
In most vertebrates, teeth are continuously shed and replaced throughout life, while mammals and several lineages of reptiles have reduced replacement to only one or two generations. In contrast to the vast majority of their living relatives, members of the lizard families Chamaeleonidae and Agamidae have dispensed with lifelong tooth replacement, instead developing acrodont dentition that fuses to the jawbone to be used for the lifetime of the animal. Though, the loss of tooth replacement has not come without a cost. In order to mitigate the consequences that come with tooth replacement loss, mammals and acrodont lizards have evolved adaptations that strengthen enamel structure and minimize wear and tear experienced during the life of the animal. While these physical adaptations are well documented, the effect that loss of tooth replacement has had on the molecular components of teeth has not received significant attention. Here, we analyze the coding and amino acid sequences of six tooth proteins (AMBN, AMEL, AMTN, ACP4, ENAM, and MMP20) from acrodont lizards, pleurodont lizards that replace teeth, and mammals. We show that the reduction of tooth generations has disproportionately affected the evolutionary trajectory of proteins associated with enamel structure, with a particularly magnified effect on the evolution of AMEL.
Keywords: Acrodont; Agamidae; Chameleon; Enamel matrix proteins; Pleurodont; Tooth replacement.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interests: The authors have no conflicts of interest to declare. All co-authors have seen and agree with the contents of the manuscript and there is no financial interest to report. We certify that the submission is original work and is not under review at any other publication.
Figures


Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Functions of secretory calcium-binding phosphoproteins in dental mineralization.J Bone Miner Res. 2025 Jul 28;40(8):909-930. doi: 10.1093/jbmr/zjaf062. J Bone Miner Res. 2025. PMID: 40318225 Review.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Continuous tooth replacement: what can teleost fish teach us?Biol Rev Camb Philos Soc. 2024 Jun;99(3):797-819. doi: 10.1111/brv.13045. Epub 2023 Dec 27. Biol Rev Camb Philos Soc. 2024. PMID: 38151229 Review.
References
-
- Abbarin N, San Miguel S, Holcroft J, Iwasaki K, Ganss B (2015) The enamel protein amelotin is a promoter of hydroxyapatite mineralization. J Bone Miner Res 30:775 - PubMed
-
- Al-Hashimi N, Lafont AG, Delgado S, Kawasaki K, Sire JY (2010) The enamelin genes in lizard, crocodile, and frog and the pseudogene in the chicken provide new insights on enamelin evolution in tetrapods. Mol Biol Evol 27:2078 - PubMed
-
- Assaraf-Weill N, Gasse B, Al-Hashimi N, Delgado S, Sire JY, Davit-Béal T (2013) Conservation of amelogenin gene expression during tetrapod evolution. Evol Dev 320:200 - PubMed
LinkOut - more resources
Full Text Sources

