Overexpression of the signaling coordinator GAB2 can play an important role in acute myeloid leukemia progression
- PMID: 40773291
- PMCID: PMC12578389
- DOI: 10.1172/JCI195929
Overexpression of the signaling coordinator GAB2 can play an important role in acute myeloid leukemia progression
Abstract
Mutations that initiate acute myeloid leukemia (AML) can cause clonal expansion without transformation (clonal hematopoiesis). Cooperating mutations, usually in signaling genes, are needed to cause overt disease, but these may require a specific fitness state to be tolerated. Here, we show that nearly all AMLs arising in a mouse model expressing 2 common AML-initiating mutations (Dnmt3aR878H and Npm1cA) acquired a single copy amplification of chromosome 7 (chr7), followed by activating mutations in signaling genes. We show that overexpression of a single gene on chr7 (Gab2, which coordinates signaling pathways) was tolerated in the presence of the Npm1cA mutation, could accelerate the development of AML, and was important for the survival of fully transformed AML cells. GAB2 is likewise overexpressed in many human AMLs with mutations in NPM1 and/or signaling genes, and also in acute promyelocytic leukemia initiated by PML::RARA; the PML::RARA fusion protein may activate GAB2 by directly binding to its 5' flanking region. A similar pattern of GAB2 overexpression preceding mutations in signaling genes has been described in other human malignancies. GAB2 overexpression may represent an oncogene-driven adaptation that facilitates the action of signaling mutations, suggesting an important (and potentially targetable) missing link between the initiating and progression mutations associated with AML.
Keywords: Genetics; Leukemias; Oncology.
Conflict of interest statement
Figures



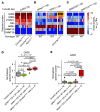



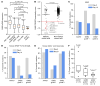

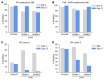
Comment in
- GAB2 couples genetic drivers and signaling networks in acute myeloid leukemia doi: 10.1172/JCI198684
References
-
- NIH. Cancer Stat Facts: Leukemia — Acute Myeloid Leukemia (AML). https://seer.cancer.gov/statfacts/html/amyl.html Accessed September 17, 2025.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

