Longitudinal single-cell analysis of glucagon-like peptide-2 treatment in patients with short bowel syndrome
- PMID: 40773292
- PMCID: PMC12487850
- DOI: 10.1172/jci.insight.194497
Longitudinal single-cell analysis of glucagon-like peptide-2 treatment in patients with short bowel syndrome
Abstract
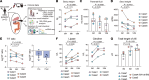
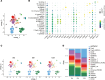
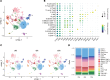
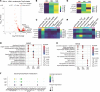
BACKGROUNDGlucagon-like peptide-2 (GLP-2) analogs are used clinically to enhance nutrient absorption in patients with short bowel syndrome (SBS); however, the precise mechanism remains unclear. To address this, the study aimed to clarify the dynamics of intestinal epithelial cells and immune cells in patients with SBS treated with GLP-2 analogs.METHODSFive male patients diagnosed with SBS, all of whom received treatment with the GLP-2 analog teduglutide, were included in the study. We conducted longitudinal single-cell RNA sequencing (scRNA-Seq) analysis of intestinal tissue from patients with SBS over a year, integrating microbiome composition analysis.RESULTSAfter treatment, the α-diversity of the gut microbiome increased, indicating a more varied microbial environment. ScRNA-Seq analysis revealed a reduction of T helper 2 cells and an increase in regulatory T cells, suggesting a shift toward an immunoregulatory intestinal environment. Additionally, nutrient-absorbing enterocyte-Top2 and middle clusters expanded, enhancing the absorption capacity, whereas major histocompatibility complex class I/II-expressing enterocyte-Top1 cells declined, potentially modulating immune responses.CONCLUSIONThe study findings indicate that GLP-2 analogs reshape intestinal immunity and microbiota, fostering a less inflammatory environment while promoting nutrient uptake efficiency. These insights offer a deeper understanding of the role of GLP-2 analogs in gut adaptation and provide a foundation for refining clinical strategies for SBS treatment.FUNDINGThis work was supported by Sakaguchi Memorial Foundation, Grants-in-Aid from the Japanese Society for the Promotion of Science (JSPS) (21K18272, 23H03665, 23H02899, 23K27590, 25K22627, 23K08037), JST FOREST(21457195), and the Takeda Japan Medical Office Funded Research Grant 2022.
Keywords: Clinical Research; Drug therapy; Gastroenterology.
Conflict of interest statement
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

