Sensitive neoantigen discovery by real-time mutanome-guided immunopeptidomics
- PMID: 40775223
- PMCID: PMC12332187
- DOI: 10.1038/s41467-025-62647-4
Sensitive neoantigen discovery by real-time mutanome-guided immunopeptidomics
Abstract
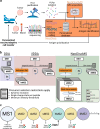
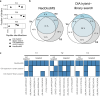
Targeting cancer-specific HLA-peptide complexes is a promising approach in immunotherapy. Mutated neoantigens are excellent targets due to their immunogenicity and cancer-specificity. Mass spectrometry (MS)-based immunopeptidomics guides the selection of naturally presented immunogenic targets within the immunopeptidome, refining immunogenicity predictions. Implementation in clinical settings, however, must achieve global depth, capturing the entirety of the immunopeptidome, maintain high target sensitivity, and cater to scarce sample inputs and short turnaround time. Here, we present NeoDiscMS, an extension of NeoDisc that enables the acquisition of personalized immunopeptidomics data. Leveraging next-generation sequencing-guided real-time spectral acquisitions, NeoDiscMS maximizes sensitivity with minimal loss of global depth. Designed for effectiveness and ease of use, with minimal effort required for implementation, NeoDiscMS enhances the detection of peptides derived from tumor-associated antigens by up to 20% and improves confidence in neoantigen identification compared to the gold standard method. NeoDiscMS advances personalization in clinical antigen discovery with more confident neoantigen detection and easy implementation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Chong, C., Coukos, G. & Bassani-Sternberg, M. Identification of tumor antigens with immunopeptidomics. Nat. Biotechnol.40, 175–188 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

