Investigating the shared genetic information between serum concentration levels of liver enzymes and cholelithiasis
- PMID: 40775300
- PMCID: PMC12330097
- DOI: 10.1186/s12876-025-04162-w
Investigating the shared genetic information between serum concentration levels of liver enzymes and cholelithiasis
Abstract
Background: Liver injury is associated with cholelithiasis, with changes in liver enzyme levels potentially influencing cholelithiasis risk. This study investigates the shared genetic basis between serum levels of four liver enzymes and cholelithiasis using summary data from large-scale genome-wide association studies (GWAS).
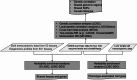
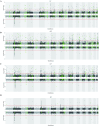
Methods: We assessed genetic correlation between liver enzymes and cholelithiasis, and performed local genetic correlation analysis to identify shared genomic regions. A cross-trait meta-analysis identified significant SNPs (single nucleotide polymorphisms) shared between the enzymes and cholelithiasis. To explore causal effects, we applied both two-sample Mendelian randomization and multivariable Mendelian randomization. Heritability-based enrichment analysis was employed to identify tissues and cells jointly associated with liver enzymes and cholelithiasis, while summary-data-based Mendelian Randomization (SMR) was utilized to identify shared genes.
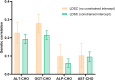
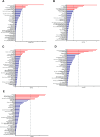
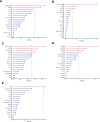
Results: Alanine aminotransferase (ALT) and gamma-glutamyl transferase (GGT) showed relatively stronger genetic correlations with cholelithiasis compared to the other liver enzymes. Shared SNPs were identified among ALT, GGT, alkaline phosphatase (ALP), and cholelithiasis. Mendelian randomization indicated that a tenfold increase in ALT could raise cholelithiasis risk by 203.4%. The liver was identified as the primary tissue linking these enzymes to cholelithiasis, but no shared cell types were implicated. Several candidate genes, such as SPTLC3, may jointly influence liver enzyme levels and cholelithiasis risk.
Conclusions: Elevated ALT levels may increase cholelithiasis risk. Genetic associations across tissues, genes, and SNPs suggest that liver enzymes could mediate the relationship between liver injury and cholelithiasis risk, providing insights into shared genetic mechanisms with potential implications for future research.
Keywords: Cholelithiasis; GWAS; Liver enzymes; Liver injury; Shared genetic information.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All data used in this study were obtained from publicly available sources and did not require ethical approval. Consent for publication: All data utilized in this study were publicly accessible and devoid of personally identifiable information; thus, informed consent was not necessary. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Serum liver enzymes and risk of stroke: Systematic review with meta-analyses and Mendelian randomization studies.Eur J Neurol. 2024 Dec;31(12):e16506. doi: 10.1111/ene.16506. Epub 2024 Oct 10. Eur J Neurol. 2024. PMID: 39387527 Free PMC article.
-
[Multi-omics Mendelian randomization study on the causality between non-ionizing radiation and facial aging].Zhonghua Shao Shang Yu Chuang Mian Xiu Fu Za Zhi. 2025 Jun 20;41(6):594-603. doi: 10.3760/cma.j.cn501225-20240830-00320. Zhonghua Shao Shang Yu Chuang Mian Xiu Fu Za Zhi. 2025. PMID: 40588408 Free PMC article. Chinese.
-
Identification of genetic association between mitochondrial dysfunction and knee osteoarthritis through integrating multi-omics: a summary data-based Mendelian randomization study.Clin Rheumatol. 2024 Nov;43(11):3487-3496. doi: 10.1007/s10067-024-07136-7. Epub 2024 Sep 11. Clin Rheumatol. 2024. PMID: 39259428 Free PMC article.
-
Statins for non-alcoholic fatty liver disease and non-alcoholic steatohepatitis.Cochrane Database Syst Rev. 2013 Dec 27;2013(12):CD008623. doi: 10.1002/14651858.CD008623.pub2. Cochrane Database Syst Rev. 2013. PMID: 24374462 Free PMC article.
-
Genetic association analysis between LDL-c lowering drugs and portal hypertension using Mendelian randomization analysis.Sci Rep. 2025 Jul 1;15(1):22238. doi: 10.1038/s41598-025-08153-5. Sci Rep. 2025. PMID: 40596422 Free PMC article.
References
-
- Wang X, Yu W, Jiang G, et al. Global epidemiology of gallstones in the 21st century: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2024;22:1586–95. - PubMed
-
- Lammert F, Gurusamy K, Ko CW, et al. Gallstones Nat Rev Dis Primers. 2016;2:16024. - PubMed
-
- Baron TH, Grimm IS, Swanstrom LL. Interventional approaches to gallbladder disease. N Engl J Med. 2015;373:357–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

