Pan-cancer landscape of ITGAV and its potential role in gastric cancer
- PMID: 40775431
- PMCID: PMC12332073
- DOI: 10.1038/s41598-025-14342-z
Pan-cancer landscape of ITGAV and its potential role in gastric cancer
Abstract
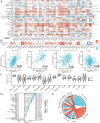
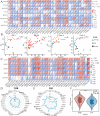
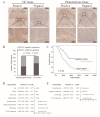
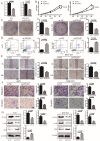
Integrin subunit alpha V (ITGAV), a subunit of the integrin receptor, is involved in many types of cancers. In order to explore the potential mechanisms of ITGAV in cancers, we carried out a comprehensive pan-cancer analysis using public database. In this study, ITGAV expression in different cancers and the relationship between ITGAV and clinic-pathological features, prognosis, genetic alteration, epigenetic modification, and tumor immune microenvironment were systemically analyzed. Gene enrichment analysis was performed to explore potential functions of ITGAV in gastric cancer (GC). GC tissue microarrays and in vitro cell experiments were used to verify the prediction results in GC. The results revealed that ITGAV was variably expression in different cancers, and ITGAV had a certain prognostic and diagnostic value in most cancers, including GC. ITGAV expression was found to be related to genetic alteration, DNA methylation, immune checkpoint gene, and immune cell infiltration in multiple cancers. Functional analyses revealed that ITGAV was involved in the regulation of EMC remodeling, ferroptosis, and cuproptosis in GC. In vitro experiments verified that ITGAV was correlated with GC cell proliferation, apoptosis, migration, and invasion. Our study demonstrated that ITGAV can be used as an effective prognostic and immunological biomarker for multiple cancers. ITGAV can promote GC malignant progression and could serve as a potential therapeutic target for GC treatment.
Keywords: Bioinformatics analysis; Biomarker; Gastric cancer; ITGAV; Pan-cancer.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval: This study was approved by the Ethics Committee of Tianjin cancer hospital (No.bc2023054) and written informed consent was obtained from all patients. All methods in this study were performed in accordance with the principles of the Declaration of Helsinki and current ethical guidelines.
Figures



Similar articles
-
Comprehensive pan-cancer analysis reveals NTN1 as an immune infiltrate risk factor and its potential prognostic value in SKCM.Sci Rep. 2025 Jan 25;15(1):3223. doi: 10.1038/s41598-025-85444-x. Sci Rep. 2025. PMID: 39863609 Free PMC article.
-
Interplay between tumor mutation burden and the tumor microenvironment predicts the prognosis of pan-cancer anti-PD-1/PD-L1 therapy.Front Immunol. 2025 Jul 24;16:1557461. doi: 10.3389/fimmu.2025.1557461. eCollection 2025. Front Immunol. 2025. PMID: 40777041 Free PMC article.
-
Comprehensive multi-omics pan-cancer analysis revealed EGFLAM as a potential prognostic and immune infiltration-associated biomarker.BMC Cancer. 2025 Jul 1;25(1):1109. doi: 10.1186/s12885-025-14519-9. BMC Cancer. 2025. PMID: 40597847 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
References
-
- Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J. Clini.74, 229–263. 10.3322/caac.21834 (2024). - PubMed
-
- Hynes, R. O. Integrins: Bidirectional, allosteric signaling machines. Cell110, 673–687. 10.1016/s0092-8674(02)00971-6 (2002). - PubMed
MeSH terms
Substances
Grants and funding
- 81401952/National Natural Science Foundation of China
- TJYXZDXK-009A/Tianjin Key Medical Discipline(Specialty) Construction Project
- TJYXZDXK-009A/Tianjin Key Medical Discipline(Specialty) Construction Project
- TJYXZDXK-009A/Tianjin Key Medical Discipline(Specialty) Construction Project
- TJYXZDXK-009A/Tianjin Key Medical Discipline(Specialty) Construction Project
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

