Proteomic Analysis of Bone Marrow CD138+ Cells to Identify Proteins Associated With the Response of Multiple Myeloma Patients to Commonly Used Therapeutic Regimens
- PMID: 40776449
- PMCID: PMC12381911
- DOI: 10.1002/pmic.70025
Proteomic Analysis of Bone Marrow CD138+ Cells to Identify Proteins Associated With the Response of Multiple Myeloma Patients to Commonly Used Therapeutic Regimens
Abstract
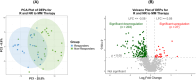
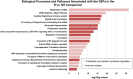
Multiple myeloma (MM) remains incurable; gaps in our understanding of MM molecular pathogenesis and drugs' resistance mechanisms are involved in the failure of therapies. This study aims to identify proteins significantly impacting MM patients' response to commonly used therapeutic regimens. Bone marrow CD138+ selected plasma cells were isolated from patients who had achieved Response (Responders, R) and those who were Non-Responders (NR) to their primary MM therapy. We used LC-MS/MS to investigate the proteomic profile of MM samples, followed by bioinformatics analysis. We identified 1190 proteins, of which 230 showed a statistically significant difference between R and NR, with 27 proteins being upregulated and 203 downregulated in R compared to NR. Pathway enrichment analysis identified pathways related to the immune response and protein synthesis regulation, closely associated with MM progression and response to therapy. The results were validated through individual RNA dataset analysis, corroborating the differential expression of several proteins, including proteins associated with MM (e.g., MIF, ILF3) as well as novel findings (e.g., DCPS and SET). Collectively, proteomics data obtained from R and NR to MM therapy displayed significant changes in the immune system and protein synthesis regulation, supporting their potential role in progression and therapeutic response of MM.
Keywords: biomarkers; cancer; multiple myeloma; proteomics; therapy.
© 2025 The Author(s). Proteomics published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

