A CD1c lipid agnostic T cell receptor bispecific engager redirects T cells against CD1c+ cells
- PMID: 40777002
- PMCID: PMC12328196
- DOI: 10.3389/fimmu.2025.1614610
A CD1c lipid agnostic T cell receptor bispecific engager redirects T cells against CD1c+ cells
Abstract
Introduction: Immunotherapy is emerging as an efficacious treatment for some cancers, complementing traditional chemo-radiation therapies. Specific markers at the cell surface of cancer cells can be used as immunotherapy targets. However, many of these markers are defined by a patient's genetic background, limiting their use across the human population.
Methods: Here, we investigated the non-polymorphic antigen presenting molecule, CD1c, that is only expressed on subsets of mature hematopoietic cells, as a potential immunotherapy target with reduced risk of off-tumor on-target toxicity in healthy tissues.
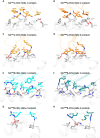
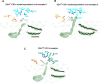
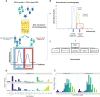
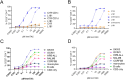
Results and discussion: We identified a T cell receptor (TCR) which recognises CD1c in a lipid independent manner and determined the crystal structure of the TCR-CD1c complex which revealed flexibility around the lipid binding region, and a new binding mechanism of auto-antigen recognition. We generated affinity enhanced variants of the TCR and fused them to an anti-CD3 antibody for T cell redirection. Lipidomic analysis revealed promiscuous lipid recognition of CD1c by the affinity enhanced TCR variants, with preference for larger lipid head group, a finding which is supported by the crystal structure. The bispecific molecule induced potent re-directed T cell killing of CD1c positive cell lines. These proof-of-concept findings demonstrate that CD1c targeting TCR bispecific engagers might be good candidates for the development of non-MHC restricted, universal therapeutics for the treatment of CD1c+ leukemias.
Keywords: CD1c; T cell engager; T cell receptor; bispecifics; immunotherapy; leukemia; lipids.
Copyright © 2025 Szoke-Kovacs, Khakoo, Rangel, Della Cristina, Pentier, Khanolkar, El-Ajouz, Simmons, Cole, Gogolak, Salio and Karuppiah.
Conflict of interest statement
RS-K, SK, RS, PD, JP, RK, SE-A, DC, VK, VR and MS are current or former employees of Immunocore LTD and may hold Immunocore stock options. PG supervised the PhD research at the University of Debrecen. The author (MS) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures

Similar articles
-
A structural perspective of how T cell receptors recognize the CD1 family of lipid antigen-presenting molecules.J Biol Chem. 2024 Aug;300(8):107511. doi: 10.1016/j.jbc.2024.107511. Epub 2024 Jun 28. J Biol Chem. 2024. PMID: 38945451 Free PMC article. Review.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Targeting of acute myeloid leukemia by five-gene engineered T cells expressing transgenic T-cell receptor specific to WT1, chimeric antigenic receptor specific to GM-CSF receptor, bispecific T-cell engager specific to CD33, and tEGFR suicide gene system.Immunother Adv. 2025 Jun 11;5(1):ltaf022. doi: 10.1093/immadv/ltaf022. eCollection 2025. Immunother Adv. 2025. PMID: 40735486 Free PMC article.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
B cell antigens: A key to optimizing CAR-T cell therapy.Int Rev Immunol. 2025 Jun 19:1-28. doi: 10.1080/08830185.2025.2515839. Online ahead of print. Int Rev Immunol. 2025. PMID: 40537997 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

