Identification of a coagulation-related classification and signature that predict disease heterogeneity for colorectal cancer and pan-cancer patients
- PMID: 40777024
- PMCID: PMC12328357
- DOI: 10.3389/fimmu.2025.1572701
Identification of a coagulation-related classification and signature that predict disease heterogeneity for colorectal cancer and pan-cancer patients
Abstract
Background: While increased coagulation is linked to cancer progression, the specific roles of coagulation-related genes in colorectal cancer (CRC) have not been extensively studied. This research identified coagulation-related subtypes (CRSs) and evaluated a coagulation-related risk score for its prognostic value in CRC.
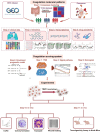
Methods: CRC dataset from The Cancer Genome Atlas was analyzed to identify CRSs using nonnegative matrix factorization, which was validated across GSE39582 and pan-cancer datasets. A list of 285 coagulation-related genes was used to develop a risk signature via least absolute shrinkage and selection operator and multivariate Cox regression. We also assessed immune characteristics and treatment responses using single-sample gene set enrichment analysis, Tumor Immune Dysfunction and Exclusion, and immunophenoscore, and constructed an overall survival-related nomogram.
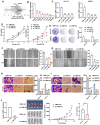
Results: CRS analysis categorized pan-cancers, including CRC, into three clusters: C1 with poor immune infiltration but better prognosis, C2 with high immune activity and prolonged survival, and C3 marked by dense immunosuppressive cells correlating with poor outcomes. Drug sensitivity analysis showed distinct responses across CRSs, influencing treatment choices. We developed a coagulation-related risk score based on F2RL2, GP1BA, MMP10, and TIMP1, which stratified CRC patients by outcome and correlated with distinct patterns of immune infiltration and therapeutic response. A validated nomogram incorporating age, TNM stage, and risk score accurately predicted overall survival, while experimental validations confirmed the bioinformatics predictions regarding TIMP1's role in CRC progression.
Conclusions: A coagulation-based classifier effectively categorizes CRC and potentially other cancers, interacting significantly with the immune microenvironment to influence disease progression and treatment responsiveness. This approach offers valuable insights for personalized cancer therapy.
Keywords: clustering; coagulation; colorectal cancer; precision treatment; tumor microenvironment.
Copyright © 2025 Pei, Gao, Xing, Chen and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures










References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

