This is a preprint.
Genetic Modulation of Lifespan: Dynamic Effects, Sex Differences, and Body Weight Trade-offs
- PMID: 40777243
- PMCID: PMC12330653
- DOI: 10.1101/2025.04.27.649857
Genetic Modulation of Lifespan: Dynamic Effects, Sex Differences, and Body Weight Trade-offs
Abstract
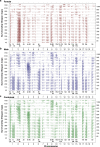
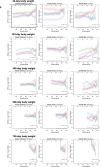
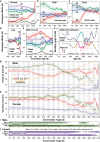
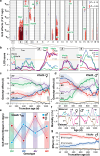
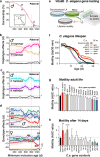
The dynamics of lifespan are shaped by DNA variants that exert effects at different ages. We have mapped genetic loci that modulate age-specific mortality using an actuarial approach. We started with an initial population of 6,438 pubescent siblings and ended with a survivorship of 559 mice that lived to at least 1100 days. Twenty-nine Vita loci dynamically modulate the mean lifespan of survivorships with strong age- and sex-specific effects. Fourteen have relatively steady effects on mortality while other loci act forcefully only early or late in life and with polarities of effects that invert. A distinct set of 19 Soma loci shape the negative correlation between weights of young adults with their life expectancies-much more strongly so in males than females. Another set of 11 Soma loci shape the positive correlation between weights at older ages with life expectancies. The Vita and Soma loci share 289 age-dependent epistatic interactions (LODs ≥3.8) but fewer than 4% are common to both sexes. We provide two examples of how to move from maps toward potential mechanisms. Our findings provide an empirical bridge between evolutionary theories on aging and genetic and molecular causes. These loci and their interactions are key to begin to understand the impact of interventions that may extend healthy lifespan in mice and even in humans.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures










References
-
- Maynard Smith J. Review lectures on senescence - I. The causes of ageing. Proc. R. Soc. Lond. B. 157, 115–127 (1962). - PubMed
-
- Rose M. & Charlesworth B. A test of evolutionary theories of senescence. Nature 287, 141–142 (1980). - PubMed
-
- Schächter F., Cohen D. & Kirkwood T. Prospects for the genetics of human longevity. Hum Genet 91, 519–526 (1993). - PubMed
Publication types
Grants and funding
- R01 AG070913/AG/NIA NIH HHS/United States
- P30 AG025707/AG/NIA NIH HHS/United States
- P30 DA044223/DA/NIDA NIH HHS/United States
- R01 GM123489/GM/NIGMS NIH HHS/United States
- R56 AG066625/AG/NIA NIH HHS/United States
- U01 AG022308/AG/NIA NIH HHS/United States
- R01 AG043930/AG/NIA NIH HHS/United States
- U01 AG022307/AG/NIA NIH HHS/United States
- R21 AG055841/AG/NIA NIH HHS/United States
- R01 AG075813/AG/NIA NIH HHS/United States
- U01 AG022303/AG/NIA NIH HHS/United States
- P30 AG024824/AG/NIA NIH HHS/United States
- P30 AG013319/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
