This is a preprint.
Calyculin A Induces Premature Chromosome Condensation and Chromatin Compaction in G1-Phase HeLa Cells without Histone H1 Phosphorylation
- PMID: 40777323
- PMCID: PMC12330644
- DOI: 10.1101/2025.07.16.665228
Calyculin A Induces Premature Chromosome Condensation and Chromatin Compaction in G1-Phase HeLa Cells without Histone H1 Phosphorylation
Abstract
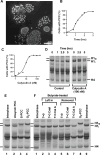
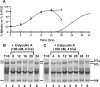
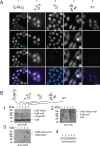
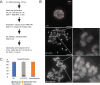
We show here that treatment of HeLa cells with calyculin A, an inhibitor of Protein Phosphatases 1 and 2A, induces premature chromosome condensation (PCC) at any point in interphase of the cell cycle. Chromosomes in G1-phase PCC closely resemble metaphase chromatids in the light microscope, and measurements using FLIM-FRET show that they have the same level of chromatin compaction as metaphase chromosomes. However, histone H1 is not phosphorylated in G1- or early S-phase PCC. These results suggest that H1 phosphorylation is not required for mitotic chromosome condensation and chromatin compaction. They also confirm that Cdk1/cyclin B, which directly phosphorylates histone H1, is not active in G1 and thus is not essential for G1-PCC. We suggest that induction of G1-PCC involves protein kinases or other factors that are either held in an inactive state by protein phosphatases, or constitutively active but countered by phosphatases. The same factors may be involved in the onset of normal mitosis, becoming active when protein phosphatases are downregulated. Induction of PCC with calyculin A should provide a useful system for identifying and studying the biochemical pathways that are required for mitotic chromosome compaction, nuclear envelope breakdown, and other events of mitosis.
Figures

Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2020 Jan 9;1(1):CD011535. doi: 10.1002/14651858.CD011535.pub3. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Apr 19;4:CD011535. doi: 10.1002/14651858.CD011535.pub4. PMID: 31917873 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
References
-
- Ajiro K, Yoda K, Utsumi K, Nishikawa Y (1996) Alteration of cell cycle-dependent histone phosphorylations by okadaic acid. Induction of mitosis-specific H3 phosphorylation and chromatin condensation in mammalian interphase cells. J Biol Chem 271:13197–201. doi: 10.1074/jbc.271.22.13197. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
