This is a preprint.
SHAPE-based chemical probes for studying preQ1-RNA interactions in living bacteria
- PMID: 40777364
- PMCID: PMC12330648
- DOI: 10.1101/2025.07.21.665968
SHAPE-based chemical probes for studying preQ1-RNA interactions in living bacteria
Update in
-
SHAPE-Based Chemical Probes for Studying preQ1-RNA Interactions in Living Bacteria.ACS Chem Biol. 2025 Nov 21;20(11):2689-2697. doi: 10.1021/acschembio.5c00548. Epub 2025 Oct 10. ACS Chem Biol. 2025. PMID: 41073353 Free PMC article.
Abstract
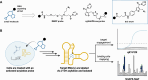
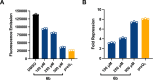
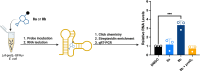
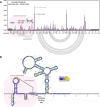
Interrogating RNA-small molecule interactions inside cells is critical for advancing RNA-targeted drug discovery. In particular, chemical probing technologies that both identify small molecule-bound RNAs and define their binding sites in the complex cellular environment will be key for establishing the on-target activity necessary for successful hit-to-lead campaigns. Using the small molecule metabolite preQ1 and its cognate riboswitch RNA as a model, herein we describe a chemical probing strategy for filling this technological gap. Building on well-established RNA acylation chemistry employed by in vivo click selective 2'-hydroxyl acylation analyzed by primer extension (icSHAPE) probes, we developed an icSHAPE-based preQ1 probe that retains biological activity in a preQ1 riboswitch reporter assay and successfully enriches the preQ1 riboswitch from living bacterial cells. Further, we map the preQ1 binding site on probe-modified riboswitch RNA by mutational profiling (MaP). As the need for rapid profiling of on- and off-target small molecule interactions continues to grow, this chemical probing strategy offers a method to interrogate cellular RNA-small molecule interactions and support the future development of RNA-targeted therapeutics.
Keywords: RNA; SHAPE; SHAPE-MaP; chemical probes; preQ1; small molecules; target engagement.
Figures

References
-
- Cao X., Zhang Y., Ding Y. & Wan Y. Probing the dynamic RNA structurome and its functions. Nat. Rev. Mol. Cell Biol. 25, 784–801 (2024). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
