This is a preprint.
snATAC-Express infers Gene Expression from Prioritized Chromatin Accessibility Peaks using Machine Learning
- PMID: 40777427
- PMCID: PMC12330625
- DOI: 10.1101/2025.07.25.666784
snATAC-Express infers Gene Expression from Prioritized Chromatin Accessibility Peaks using Machine Learning
Abstract
Background: Single cell multi-omic investigation opens-up new opportunities to understand mechanisms of gene regulation. Existing methods for inferring transcript abundance from chromatin accessibility fail to prioritize the most relevant peaks and tend to assume positive associations between ATAC peaks and RNA counts. We hypothesize that gene regulation can be modeled as a function of combined positive and negative interactions among peaks and that causal regulatory variants are enriched in the vicinity of the most critical peaks.
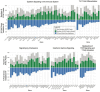
Results: A machine learning pipeline leveraging single nuclear multiomic transcriptome and chromatin accessibility data is developed to model gene expression as a function of ATAC peak intensity. Multiome data was available for 18 immune cell types from 29 donors, 19 with Crohn's disease. The pipeline aggregates results from three machine learning approaches (random forest regression, XGBoost, and Light GBM) as well as linear regression to identify which ATAC peaks contribute to explaining variation among donors and cell types in pseudobulk gene expression. The coefficient of determination with cross-validation was used to identify robust models which typically explain between 5% and 40% of transcript abundance, utilizing on average 47% of the ATAC peaks, representing a significant gain in predictive accuracy. The most important peaks are enriched in GWAS variants for inflammatory bowel disease and the autoimmune disease systemic lupus erythematosus, but not for rheumatoid arthritis.
Conclusion: Atlanta Plots visualize the proportion of ATAC peaks contributing to a predictive model of gene expression as well as the proportion of variance explained by the model. Software implementing our pipeline, "snATAC-Express", is freely available on GitHub.
Keywords: GWAS enrichment; Gene regulation; chromatin accessibility; machine learning; multi-omics.
Conflict of interest statement
Competing Interests: The authors declare that they have no competing interests. RDW is the developer of JMP commercial statistical software that includes machine learning tools similar to those described here, but all code was and can be implemented with open source software.
Figures





Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Comprehensive single-cell chromatin and transcriptomic profiling of peripheral immune cells in nonsegmental vitiligo.Br J Dermatol. 2025 Jun 20;193(1):115-124. doi: 10.1093/bjd/ljaf041. Br J Dermatol. 2025. PMID: 39888372
-
An optimized approach for multiplexing single-nuclear ATAC-seq using oligonucleotide-conjugated antibodies.Epigenetics Chromatin. 2023 Apr 28;16(1):14. doi: 10.1186/s13072-023-00486-7. Epigenetics Chromatin. 2023. PMID: 37118773 Free PMC article.
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
