This is a preprint.
Chemically modified CRISPR-Cas9 enables targeting of individual G-quadruplex and i-motif structures, revealing ligand-dependent transcriptional perturbation
- PMID: 40777503
- PMCID: PMC12330486
- DOI: 10.1101/2024.10.14.618195
Chemically modified CRISPR-Cas9 enables targeting of individual G-quadruplex and i-motif structures, revealing ligand-dependent transcriptional perturbation
Abstract
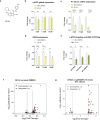
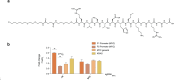
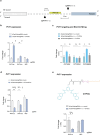
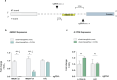
The development of selective ligands to target DNA G-quadruplexes (G4s) and i-motifs (iMs) has revealed their relevance in transcriptional regulation. However, most of these ligands are unable to target individual G4s or iMs in the genome, severely limiting their scope. Herein, we describe a new Approach to Target Exact Nucleic Acid alternative structures (ATENA) that relies on the chemical conjugation of established G4 and iM ligands to a catalytically inactive Cas9 protein (dCas9), enabling their individual targeting in living cells. ATENA demonstrated that the selective targeting of the G4 present in the oncogene c-MYC leads to the suppression of transcripts regulated exclusively by one of its promoters (P1). Conversely, targeting the c-MYC iMs on the opposite strand leads to the selective increase of P1-driven transcripts. ATENA revealed that G4-mediated transcriptional responses are highly ligand-specific, with different ligands eliciting markedly different effects at the same G4-site. We further demonstrated that the basal expression levels of the gene targeted can be used to predict the transcriptional impact associated with G4-stabilization. Our study provides an innovative platform to investigate G4- and iM-biology with high precision and unveils the therapeutic relevance of individual DNA structures with unprecedented selectivity.
Figures


References
-
- Davis J. T. G-quartets 40 years later: from 5’-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 43, 668–698 (2004). - PubMed
-
- Parkinson G. N., Lee M. P. & Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417, 876–880 (2002). - PubMed
Publication types
LinkOut - more resources
Full Text Sources
