Off-target effects in CRISPR-Cas genome editing for human therapeutics: Progress and challenges
- PMID: 40777742
- PMCID: PMC12329535
- DOI: 10.1016/j.omtn.2025.102636
Off-target effects in CRISPR-Cas genome editing for human therapeutics: Progress and challenges
Abstract
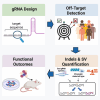
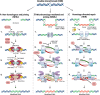
Targeted nucleases, primarily CRISPR-Cas-based systems, have revolutionized genome editing by enabling precise modification of target genes or transcripts. Many pre-clinical and clinical studies leverage this technology to develop treatments for human diseases; however, substantial off-target genotoxicity concerns delay its clinical translation. Despite the development of a wide array of tools, assays, and technologies aimed at identifying and quantifying off-target effects, the absence of standardized guidelines leads to inconsistent practices across studies. This review highlights the key challenges and potential solutions in ensuring the safety of gene editing studies for therapeutic applications, focusing on gRNA design, off-target sites prediction, and off-target activity measurement.
Keywords: CRISPR-Cas9; MT: Clinical Applications; gene therapy; genome editing; off-target; safety.
© 2025 The Authors.
Conflict of interest statement
A.H. is the founder and chief scientific officer of CassidyBio. CassidyBio did not have input into the design, execution, interpretation, or publication of this work. T.C. and G.T. hold patents on CAST-seq. G.T. is current employees of AstraZeneca and may be AstraZeneca shareholders.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources

