Obox1 deficiency impairs fertility in female mice
- PMID: 40777786
- PMCID: PMC12327860
- DOI: 10.1016/j.fmre.2025.04.008
Obox1 deficiency impairs fertility in female mice
Abstract
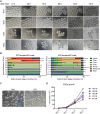
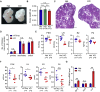
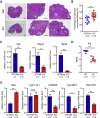
OBOX1 is a maternal factor involved in oogenesis and follicle development, yet its specific role remains unclear. Here, we demonstrated that Obox1 knockout female mice exhibit subfertility, characterized by reduced litter size and impaired ovulation. These oocytes show minimal disruption in early embryonic development post-fertilization. However, Obox1 deficiency leads to decreased levels of gonadotropins and female sex hormones, especially the luteinizing hormone (LH). Exogenous human chorionic gonadotropin (hCG) administration during superovulation failed to rescue the ovulation defect. Post-ovulation, the ovulation-related genes and serum progesterone levels were significantly reduced in Obox1-deficienct ovaries, accompanied by dysregulated steroidogenesis-related gene expression. Transcriptomic profiling of Obox1 deficient metaphase II (MII) oocytes revealed downregulation of genes involved in mitochondrial energy metabolism and biosynthesis, and upregulation of genes associated with cell transport, transcription, RNA processing, translation. Further investigation revealed that follistatin gene expression was upregulated in both MII oocytes and ovaries of Obox1 deficient mice, along with increased expression of Gdf9, Bmp15, Foxl2, and NOTCH signaling components. These findings suggest that Obox1 is essential for maintaining hormonal balance and ovulatory function through regulating oocyte-granulosa cell interactions and steroid hormone synthesis.
Keywords: Aberrant steroidogenesis; Abnormal luteinization; Hormone deficiency; Oocyte-granulosa cell interactions; Subfertility.
© 2025 The Authors. Publishing Services by Elsevier B.V. on behalf of KeAi Communications Co. Ltd.
Conflict of interest statement
The authors declare that they have no conflicts of interest in this work.
Figures

Similar articles
-
Single-cell proteomics analysis of human oocytes during GV-to-MI transition.Hum Reprod. 2025 Jul 1;40(7):1332-1343. doi: 10.1093/humrep/deaf086. Hum Reprod. 2025. PMID: 40359387
-
Rescue in vitro maturation of germinal vesicle oocytes after ovarian stimulation: the importance of the culture media.Hum Reprod. 2025 Aug 1;40(8):1504-1515. doi: 10.1093/humrep/deaf099. Hum Reprod. 2025. PMID: 40447125 Free PMC article.
-
The inhibition of prostaglandin production during ovulation by exposure to a phthalate mixture is circumvented by cAMP analogue supplementation in a human granulosa cell model.Reprod Toxicol. 2025 Sep;136:108968. doi: 10.1016/j.reprotox.2025.108968. Epub 2025 May 31. Reprod Toxicol. 2025. PMID: 40456493
-
Individualised gonadotropin dose selection using markers of ovarian reserve for women undergoing in vitro fertilisation plus intracytoplasmic sperm injection (IVF/ICSI).Cochrane Database Syst Rev. 2018 Feb 1;2(2):CD012693. doi: 10.1002/14651858.CD012693.pub2. Cochrane Database Syst Rev. 2018. Update in: Cochrane Database Syst Rev. 2024 Jan 4;1:CD012693. doi: 10.1002/14651858.CD012693.pub3. PMID: 29388198 Free PMC article. Updated.
-
Gonadotropin-releasing hormone agonist versus HCG for oocyte triggering in antagonist-assisted reproductive technology.Cochrane Database Syst Rev. 2014 Oct 31;2014(10):CD008046. doi: 10.1002/14651858.CD008046.pub4. Cochrane Database Syst Rev. 2014. PMID: 25358904 Free PMC article.
References
-
- Magalhaes R.S., Atala A. In: Principles of Regenerative Medicine. 3rd Edition. Atala A., Lanza R., Mikos A.G., et al., editors. Academic Press; Location: 2019. Chapter 70 - Regenerative Medicine for the Female Reproductive System of chapter; pp. 1237–1250.
-
- Das P.K., Mukherjee J., Banerjee D., et al. In: Textbook of Veterinary Physiology. Das P.K., Sejian V., Mukherjee J., et al., editors. Springer Nature Singapore; Location: 2023. Female Reproductive Physiology of chapter; pp. 513–568.
-
- Messinis I.E., Messini C.I., Dafopoulos K. The role of gonadotropins in the follicular phase. Ann NY Acad. Sci. 2010;1205:5–11. - PubMed
-
- Gingold J.A., Jain M., Jalai C. In: Clinical Reproductive Medicine and Surgery: A Practical Guide. Falcone T., Hurd W.W., editors. Springer Nature Switzerland AG; Cham, Switzerland: 2022. Hypothalamic-Pituitary-Ovarian Axis and Control of the Menstrual Cycle of chapter; pp. 1–22.
LinkOut - more resources
Full Text Sources

