This is a preprint.
Effective Aerosol Inoculation of Dose-Escalated Seasonal Influenza H3N2 Virus in Controlled Human Infection Model
- PMID: 40778160
- PMCID: PMC12330413
- DOI: 10.1101/2025.07.23.25332064
Effective Aerosol Inoculation of Dose-Escalated Seasonal Influenza H3N2 Virus in Controlled Human Infection Model
Abstract
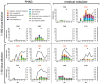
Human challenge models (CHIMs) are instrumental in advancing influenza research but have traditionally relied on intranasal inoculation, which does not mimic the natural aerosol transmission of the virus. We conducted a dose-escalation influenza CHIM study to evaluate the safety and feasibility of two modern aerosol delivery systems: a flow-focusing monodisperse aerosol generator (FMAG) and a medical nebulizer. Fourteen healthy adults aged 18-49 years were exposed to influenza A/Perth/16/2009 (H3N2) in a controlled inpatient setting. Infection rates were 75% (3/4) with FMAG and 50% (2/4) with the nebulizer at the higher dose. Infections were self-limited, with sinus congestion, rhinorrhea, and cough being the most common symptoms. No serious adverse events occurred. Viral shedding was reproducible across respiratory sites, and seroconversion occurred in 33% of infected participants. Symptom timing and viral kinetics were comparable to those observed in prior intranasal CHIMs. Participants receiving nebulizer-delivered virus showed earlier viral detection in the oral cavity, suggesting broader airway deposition. These findings demonstrate that aerosol influenza challenge is both safe and effective and can simulate natural infection more accurately than intranasal delivery. This reintroduction of aerosolized influenza challenge provides a robust platform for studying transmission dynamics, tissue-specific immune responses, and for evaluating next-generation vaccines and therapeutics under conditions that better approximate real-world exposure.
Conflict of interest statement
NR: Emory University receives grants and contracts supporting NR’s research from Merck, Sanofi, Pfizer, Vaccine Company, Immorna, and the NIH; NR has received honoraria from Virology Education and Medscape and travel support from Sanofi and Moderna; she serves on advisory boards for Moderna, Sanofi, Seqirus, and Pfizer and on safety committees for EMMES, ICON, BARDA, CyanVac, Imunon, and Micron; NR holds leadership roles on the ARLG and CDC Pertussis Challenge advisory boards, the UAB Immunology Institute advisory board, and the DMID Safety Monitoring Committee for a malaria challenge model; she has also received equipment and other support from the Georgia Research Alliance. AM and AC are employees of hVIVO and hold stock in the company. ME serves as a paid consultant to Medtronic and Boston Scientific and has received advisory-board payments for a Medtronic patent committee. CK receives CTSA, CFAR, and industry trial support to her institution; she consults for Rebiotix/Ferring (payments to her personally) and has served as an expert witness for Womble Dickinson (payments to her); she has received travel support to ASM meetings; holds a pending patent on a fecal-microbiota processor; serves unpaid on the boards of Project Mercy and the National MPS Society; and has received equipment from the Georgia Research Alliance. SL receives core laboratory support from the Flu Lab; has received honoraria from NIH, Indiana University, Yale, University of Colorado, Cleveland Clinic, Boston University, NYU, and the University of Wisconsin–Madison; and serves on the APPA Medical Advisory Board. LM: Flu Lab support to her institution; grants from NSF and Wellcome Leap (to institution); consulting fees from MITRE (paid to her); honoraria from The New York Times, University of Hong Kong, Queensland University, and Washington University (paid to her); travel support from MITRE and ISIRV; and a pending patent on a surface-enhanced Raman spectroscopy biosensor. ALow: Flu Lab support to Emory University; multiple NIAID grants (U01 AI144673, R01 AI146260, R01 AI154894, R01 AI165644, R01 AI127799, P01 AI186819, 75N93021C00017) paid to Emory; and equipment from the Georgia Research Alliance.. AJC, AL, AKM, SDB, CR, DG, KP, EF, GQ, JN, KB, KBush, JT, JD, JP, MP, MV, NS, RT, MS, and NVM report no relationships, activities, or interests to disclose.
Figures

References
-
- Smorodintseff A, Tushinsky M, Drobyshevskaya A, Korovin A, Osetroff A. Investigation on volunteers infected with the influenza virus. Am J Med Sci. 1937;194:159–70.
-
- Henle W, Henle G, Stokes J. Demonstration of the efficacy of vaccination against influenza Type A by experimental infection of human beings. J Immunol. 1943;46(3):163–75. doi: 10.4049/jimmunol.46.3.163 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
