Integrative machine learning approach for forecasting lung cancer chemosensitivity: From algorithm to cell line validation
- PMID: 40778317
- PMCID: PMC12329548
- DOI: 10.1016/j.csbj.2025.07.043
Integrative machine learning approach for forecasting lung cancer chemosensitivity: From algorithm to cell line validation
Abstract
Background: Chemotherapy remains the primary treatment modality for patients with lung cancer; however, substantial inter-patient variability exists in responses to chemotherapeutic agents. Therefore, predicting individual responses is critical for optimizing treatment outcomes and improving patient prognosis.
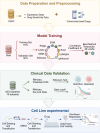
Methods: This study developed a model to predict chemotherapy response in lung cancer patients by integrating multi-omics and clinical data from the Genomics of Drug Sensitivity in Cancer database, employing 45 machine learning algorithms. Data from the Gene Expression Omnibus database were utilized to validate the model. The impact of key genes on chemotherapy response was assessed in cell lines.
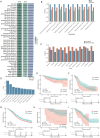
Results: A model combining random forest and support vector machine algorithms exhibited superior performance in both the training and validation sets. Furthermore, patients in the sensitive group demonstrated longer overall survival compared to those in the resistant group. TMED4 and DYNLRB1 genes were identified as pivotal features in the model and exhibited higher expression levels in the chemotherapy-resistant group. SiRNA-mediated knockdown of gene expression enhanced the chemosensitivity of lung cancer cell lines to chemotherapeutic agents.
Conclusions: This study successfully developed a high-performance machine learning model for predicting chemotherapy response in lung cancer and elucidated a strong correlation between TMED4 and DYNLRB1 gene expression and chemotherapy resistance. We further provide a user-friendly web server (available at https://smuonco.shinyapps.io/LC-DrugPortal/) to enable clinical utilization of our model, promoting personalized chemotherapy selection for lung cancer patients.
Keywords: Cell Line Validation; Chemosensitivity; Lung Cancer; Machine Learning; Prediction.
© 2025 The Authors. Published by Elsevier B.V. on behalf of Research Network of Computational and Structural Biotechnology.
Conflict of interest statement
All authors declare no competing financial or non-financial interests.
Figures



References
-
- Travis W.D., Brambilla E., Nicholson A.G., et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–1260. - PubMed
LinkOut - more resources
Full Text Sources
