Dominant-negative isoform of TDP-43 is regulated by ALS-linked RNA-binding proteins
- PMID: 40778857
- PMCID: PMC12333503
- DOI: 10.1083/jcb.202406097
Dominant-negative isoform of TDP-43 is regulated by ALS-linked RNA-binding proteins
Abstract
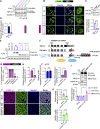
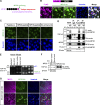
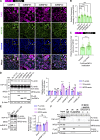
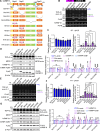
TDP-43, an RNA-binding protein (RBP) encoded by the TARDBP gene, is crucial for understanding the pathogenesis of neurodegenerative diseases like amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration. Dysregulated TDP-43 causes motor neuron loss, highlighting the need for proper expression levels. Here, we identify a dominant-negative isoform among the multiple TARDBP splicing variants and validate its endogenous expression using a developed antibody against its translated product. Furthermore, we revealed that ALS-associated RBPs regulate its expression: hnRNP K promotes its splicing and expression, while hnRNP A1 and FUS suppress these processes through distinct mechanisms. hnRNP A1 inhibits hnRNP K-mediated splicing, and FUS represses the dominant-negative isoform through both its translational inhibition and hnRNP K suppression. Notably, ALS-mutant FUS weakens this regulatory mechanism, leading to impaired repression of hnRNP K and the dominant-negative isoform. Our findings suggest a regulatory network involving ALS-linked RBPs that govern TDP-43 isoform expression and provide new insights into how disruptions in this network contribute to ALS pathogenesis.
© 2025 Hasegawa-Ogawa et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures










Similar articles
-
Dysregulated FOXO1 activity drives skeletal muscle intrinsic dysfunction in amyotrophic lateral sclerosis.Acta Neuropathol. 2024 Sep 16;148(1):43. doi: 10.1007/s00401-024-02794-y. Acta Neuropathol. 2024. PMID: 39283487 Free PMC article.
-
Regulation of physiological and pathological condensates by molecular chaperones.FEBS J. 2025 Jul;292(13):3271-3297. doi: 10.1111/febs.17390. Epub 2025 Jan 5. FEBS J. 2025. PMID: 39756021 Free PMC article. Review.
-
Altered mRNA transport and local translation in i3Neurons with RNA-binding protein knockdown.Nucleic Acids Res. 2025 Jul 19;53(14):gkaf709. doi: 10.1093/nar/gkaf709. Nucleic Acids Res. 2025. PMID: 40737092 Free PMC article.
-
Deep learning analyses of splicing variants identify the link of PCP4 with amyotrophic lateral sclerosis.Brain. 2025 Jul 7;148(7):2331-2347. doi: 10.1093/brain/awaf025. Brain. 2025. PMID: 39852553
-
[Elucidation of the Molecular Mechanism Underlying Aberrant Formation of RNA Granules in Neurons of ALS Patients and Its Regulation].Yakugaku Zasshi. 2025;145(7):583-588. doi: 10.1248/yakushi.24-00209-1. Yakugaku Zasshi. 2025. PMID: 40603049 Review. Japanese.
References
-
- Afroz, T., Hock E.M., Ernst P., Foglieni C., Jambeau M., Gilhespy L.A.B., Laferriere F., Maniecka Z., Plückthun A., Mittl P., et al. 2017. Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat. Commun. 8:45. 10.1038/s41467-017-00062-0 - DOI - PMC - PubMed
-
- An, H., Skelt L., Notaro A., Highley J.R., Fox A.H., La Bella V., Buchman V.L., and Shelkovnikova T.A.. 2019. ALS-linked FUS mutations confer loss and gain of function in the nucleus by promoting excessive formation of dysfunctional paraspeckles. Acta Neuropathol. Commun. 7:7. 10.1186/s40478-019-0658-x - DOI - PMC - PubMed
-
- Arai, T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., and Oda T.. 2006. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351:602–611. 10.1016/j.bbrc.2006.10.093 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

