Emodin Alleviates Sepsis-Induced Multiorgan Damage by Inhibiting NETosis through Targeting Neutrophils BCL-10
- PMID: 40779122
- PMCID: PMC12591137
- DOI: 10.1002/advs.202417129
Emodin Alleviates Sepsis-Induced Multiorgan Damage by Inhibiting NETosis through Targeting Neutrophils BCL-10
Abstract
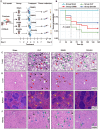
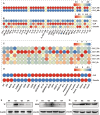
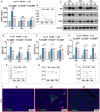
Sepsis is a life-threatening condition caused by dysregulated host responses to infection, characterized by excessive inflammation and abnormal coagulation. Neutrophil extracellular traps (NETs) formation bridges these two pathological processes. Through both in vivo and in vitro experiments, it is observed that Emodin, a natural anthraquinone derivative derived from Dahuang, significantly ameliorates the cytokine storm and coagulation abnormalities induced by sepsis, demonstrating remarkable efficacy in inhibiting NETs formation. Furthermore, through protein microarrays, surface plasmon resonance (SPR), pull-down assays, and molecular docking analyses, BCL-10 is established as a direct target of Emodin, providing protective effects in both in vivo and in vitro settings. Through conditional knockout of BCL-10 in neutrophils, alongside single-cell sequencing analyses, it is confirmed that BCL-10 is key in promoting excessive NET formation in sepsis. Additionally, Emodin exerts powerful protective effects by modulating the function of the BCL-10/MALT1 complex, thereby alleviating the NF-κB signaling activation and inhibiting NETs formation. Collectively, these findings provide pharmacological evidence that Emodin targeted BCL-10 regulates the BCL-10/MALT1 complex and suppresses NF-κB activation, ultimately conferring significant multiorgan protective effects in sepsis. The conduct of this study provides new clues for the translational research of Emodin and its target BCL-10 in sepsis.
Keywords: BCL‐10/MALT1; Emodin; NETosis; NF‐κB; sepsis.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Cecconi M., Evans L., Levy M., Rhodes A., Lancet 2018, 392, 75. - PubMed
-
- Fleischmann C., Scherag A., Adhikari N. K. J., Hartog C. S., Tsaganos T., Schlattmann P., Angus D. C., Reinhart K., Am. J. Respir. Crit. Care Med. 2016, 259. - PubMed
-
- Liu V., Escobar G. J., Greene J. D., Soule J., Whippy A., Angus D. C., Iwashyna T. J., JAMA, J. Am. Med. Assoc. 2014, 312, 90. - PubMed
-
- Hotchkiss R. S., Karl I. E., N. Engl. J. Med. 2003, 348, 138. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
