In Silico identification and modelling of FDA-approved drugs targeting T-type calcium channels
- PMID: 40779495
- PMCID: PMC12334052
- DOI: 10.1371/journal.pone.0327386
In Silico identification and modelling of FDA-approved drugs targeting T-type calcium channels
Abstract
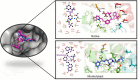
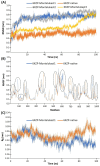
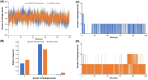
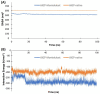
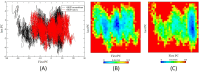
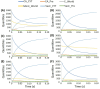
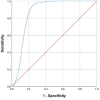
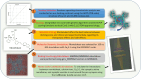
Studies have shown that inhibition of the Cav3.1 T-type calcium channel can prevent or suppress neurological diseases, such as epileptic seizures and diabetic neuropathy. In this study, we aimed to use in silico simulations to identify a U.S. Food and Drug Administration (FDA)-approved drug that can bind to the Cav3.1 T-type calcium channel. We used the automated docking suite GOLD v5.5 with the genetic algorithm to simulate molecular docking and predict the protein-ligand binding modes, and the ChemPLP empirical scoring function to estimate the binding affinities of 2,115 FDA-approved drugs to the human Cav3.1 channel. Drugs with high binding affinity and appropriate pharmacodynamic and pharmacokinetic properties were selected for molecular mechanics Poisson-Boltzmann surface area (MMPBSA) and molecular mechanics generalised Born surface area (MMGBSA) binding free energy calculations, GROMACS molecular dynamics (MD) simulations and Monte Carlo Cell (MCell) simulations. The docking results indicated that the FDA-approved drug montelukast has a high binding affinity to Cav3.1, and data from the literature suggested that montelukast has the appropriate drug-like properties to cross the human blood-brain barrier and reach synapses in the central nervous system. MMPBSA, MMGBSA, and MD simulations showed the high stability of the montelukast-Cav3.1 complex. MCell simulations indicated that the blockage of Cav3.1 by montelukast reduced the number of synaptic vesicles being released from the pre-synaptic region to the synaptic cleft, which may reduce the probability and amplitude of postsynaptic potentials.
Copyright: © 2025 Fong et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Belardetti F, Zamponi GW. Calcium channels as therapeutic targets. WIREs Membr Transp Signal. 2012;1(4):433–51. doi: 10.1002/wmts.38 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

