Precision-cut tumor tissue slices, a novel tool to study the tumor microenvironment interactions with chimeric antigen receptor (CAR) T cells
- PMID: 40779497
- PMCID: PMC12334022
- DOI: 10.1371/journal.pone.0327322
Precision-cut tumor tissue slices, a novel tool to study the tumor microenvironment interactions with chimeric antigen receptor (CAR) T cells
Abstract
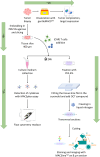
Up until present day, chimeric antigen receptor (CAR)-T cell therapy has only been approved for hematological malignancies, as CAR-T cells do not show comparable efficacy in solid tumors. Therefore, understanding the features of the tumor microenvironment (TME), is key to improve efficacy of adoptive cell therapies (ACTs) against solid tumors. In this context, robust workflows, which dissect the complex interactions between CAR-T cells and the TME are still lacking. To address this need, we have established an ex vivo workflow co-culturing tissue slices from patient tumor resections with CAR-T cells. The workflow is composed of assessing several complementary attributes, such as cytokine release via flow cytometry, quantification of cell infiltration into the tumor and assessment of the regions of the tissue slice the CAR-T cell infiltrate into by using the MACSima™ imaging cyclic staining technology. Using this workflow it is possible to observe the behavior of CAR-T cells within the tumor and its TME, their infiltration into distinct tumor compartments, as well as to dissect the underlying molecular mechanisms that drive T cell migration, thanks to MACSima™ multiplexing technology and its ability to image several markers at the same time. Assessment of ovarian carcinoma tissue slices revealed substantial release of specific cytokines and increased infiltration of T cells in the tumor areas when CAR-T cells were added to the tissue slices as compared to non-engineered T cells. The establishment of this novel approach will enable researchers to better characterize the interaction between CAR-T cells and the TME. Tissue slices present an intrinsic heterogeneity, which is indeed an advantage compared to other in vitro models but can turn itself into complex results interpretation. Therefore, we recommend that any conclusion derived from this assay should be verified with complementary models.
Copyright: © 2025 Durante et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
Valeria Durante, Alina Wittwer, Benjamin Theek, Manuel Martinez-Osuna, Olaf Hardt, Dominik Eckardt, Andreas Bosio, Sonja Schallenberg and Christoph Herbel are or were employees of Miltenyi Biotec B.V. & Co. KG. This does not alter our adherence to PLOS ONE policies on sharing data and materials
Figures




Similar articles
-
Metabolic reprogramming via an engineered PGC-1α improves human chimeric antigen receptor T-cell therapy against solid tumors.J Immunother Cancer. 2023 Mar;11(3):e006522. doi: 10.1136/jitc-2022-006522. J Immunother Cancer. 2023. PMID: 36914208 Free PMC article.
-
From spheroids to organoids: next-generation models for CAR-T cell therapy research in solid tumors.Front Immunol. 2025 Jul 11;16:1626369. doi: 10.3389/fimmu.2025.1626369. eCollection 2025. Front Immunol. 2025. PMID: 40718488 Free PMC article. Review.
-
NK Cells Expressing a Chimeric Activating Receptor Eliminate MDSCs and Rescue Impaired CAR-T Cell Activity against Solid Tumors.Cancer Immunol Res. 2019 Mar;7(3):363-375. doi: 10.1158/2326-6066.CIR-18-0572. Epub 2019 Jan 16. Cancer Immunol Res. 2019. PMID: 30651290 Free PMC article.
-
A new sort of cells for chimeric antigen receptor T-cell therapies-isolating CD14-CD127+ T cells for chimeric antigen receptor T-cell manufacture.Cytotherapy. 2025 Aug;27(8):980-990. doi: 10.1016/j.jcyt.2025.04.068. Epub 2025 Apr 18. Cytotherapy. 2025. PMID: 40380956
-
CAR-T Cells Therapy in Glioblastoma: A Systematic Review on Molecular Targets and Treatment Strategies.Int J Mol Sci. 2024 Jun 29;25(13):7174. doi: 10.3390/ijms25137174. Int J Mol Sci. 2024. PMID: 39000281 Free PMC article.
References
-
- Ledford H. Engineered cell therapy for cancer gets thumbs up from FDA advisers. Nature. 2017;547(7663):270. - PubMed
-
- Kochenderfer JN, Wilson WH, Janik JE, Dudley ME, Stetler-Stevenson M, Feldman SA, et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood. 2010;116(20):4099–102. doi: 10.1182/blood-2010-04-281931 - DOI - PMC - PubMed
-
- Administration USFD. FDA approves tisagenlecleucel for B-cell ALL and tocilizumab for cytokine release syndrome. 2017.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

