A human NK cell progenitor that originates in the thymus and generates KIR+NKG2A- NK cells
- PMID: 40779632
- PMCID: PMC12333686
- DOI: 10.1126/sciadv.adv9650
A human NK cell progenitor that originates in the thymus and generates KIR+NKG2A- NK cells
Abstract
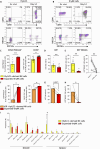
KIR+NKG2A- natural killer (NK) cells have the unique ability to detect down-regulation of single HLA-I allotypes, frequently occurring in malignantly transformed and virus-infected cells. We have recently shown that circulating innate lymphoid cells 1 (cILC1s) have the potential to generate such KIR+NKG2A- NK cells, but their developmental origin was unknown. Here, we demonstrate that the development of cILC1 is thymus dependent and identify a putative progenitor of cILC1s in the thymus (thyILC1). Single-cell RNA sequencing analysis revealed a close relationship of thyILC1s to CD34+ double-negative thymocytes. Both generated comparable NK cell frequencies, while only thyILC1s could be efficiently differentiated into KIR+NKG2A- NK cells. Last, patients with FOXN1 haploinsufficiency, showing congenital thymic hypoplasia, exhibited a profound deficiency of cILC1s but not cILC2s and cILC3s, demonstrating their specific thymus dependency. Together, the data suggest that thyILC1s are the source of a thymus-dependent NK cell differentiation pathway that promotes generation of KIR+NKG2A- NK cells.
Figures







References
-
- Hazenberg M. D., Spits H., Human innate lymphoid cells. Blood 124, 700–709 (2014). - PubMed
-
- Vivier E., Artis D., Colonna M., Diefenbach A., Di Santo J. P., Eberl G., Koyasu S., Locksley R. M., McKenzie A. N. J., Mebius R. E., Powrie F., Spits H., Innate lymphoid cells: 10 Years on. Cell 174, 1054–1066 (2018). - PubMed
-
- Bennstein S. B., Uhrberg M., Circulating innate lymphoid cells (cILCs): Unconventional lymphocytes with hidden talents. J. Allergy Clin. Immunol. 154, 523–536 (2024). - PubMed
-
- Mjösberg J. M., Trifari S., Crellin N. K., Peters C. P., van Drunen C. M., Piet B., Fokkens W. J., Cupedo T., Spits H., Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 12, 1055–1062 (2011). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

