Experimental and computational approaches for evaluating molecule interactions with equilibrative nucleoside transporters 1 and 2
- PMID: 40779912
- PMCID: PMC12486301
- DOI: 10.1016/j.jpet.2025.103660
Experimental and computational approaches for evaluating molecule interactions with equilibrative nucleoside transporters 1 and 2
Abstract
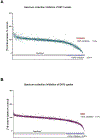
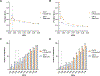
Equilibrative nucleoside transporters (ENTs) facilitate the equilibrative movement of nucleosides and nucleobases across cell membranes in a sodium-independent manner. ENT1 (SLC29A1) and ENT2 (SLC29A2) also transport nucleoside analogs and can affect the pharmacokinetics and pharmacodynamics of drugs used in cancer, viral infections, and inflammatory disorders. ENT1 and ENT2 may be differentiated functionally by their sensitivity to inhibition by nitrobenzylthioinosine (NBMPR), and we used this difference in NBMPR sensitivity to create a HeLa-based ENT2 inhibition assay. We then screened a library of 1600 diverse compounds composed of drugs and natural products for inhibition against ENT1 and ENT2, selecting a subset of compounds for side-by-side comparison of dose-response studies. We used these screening data to build machine learning models for ENT1 and ENT2 inhibition, employing dataset balancing and conformal prediction to adjust for the asymmetrical nature of the data. A random forest model predicted a prospective test set of 44 additional molecules (from the MedChem Express Drug Repurposing Library [2700 compounds]) as potential ENT1 inhibitors with 59% accuracy. This resulted in the identification of the Food and Drug Administration-approved drugs isradipine, avanafil, and istradefylline as inhibitors of ENT1. These new experimental and computational methods and models for these clinically relevant transporters can be used to evaluate drug-transporter interactions early in drug discovery, before testing in vivo. SIGNIFICANCE STATEMENT: Recent regulatory guidance have suggest the inclusion of the equilibrative nucleoside transporters (eg, ENT1 and ENT2) as transporters with emerging clinical relevance for in vitro and in vivo assessment. We have screened over 1600 diverse molecules, allowing us to build machine learning models that in turn were further used to make predictions to validate the models. Our combined experimental and machine learning approach resulted in the identification of multiple Food and Drug Administration-approved medications as inhibitors of ENT1 or ENT2.
Keywords: Compound library; Conformal prediction; Drug screening; Equilibrative nucleoside transporters; Machine learning models; Random forest model.
Copyright © 2025 American Society for Pharmacology and Experimental Therapeutics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest SE is the founder and owner of Collaborations Pharmaceuticals, Inc. PAV, JSH, RR, and TRL are employees. All other authors have no conflicts of interest.
Figures
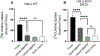



References
-
- RDKit: Open-source cheminformatics, in.
-
- Baskin II, Winkler D and Tetko IV (2016) A renaissance of neural networks in drug discovery. Expert Opin Drug Discov 11:785–795. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

